DNA-directed Synthesis of Generation 7 and 5 Polyamidoamine (PAMAM) dendrimer Nanoclusters
1Department of Biomedical Engineering, School of Engineering, University of Michigan at Ann Arbor,
Ann Arbor, MI 48109 USA
2Department of Physics, School of Literature, Art and Science, University of Michigan at Ann Arbor,
Ann Arbor, MI 48109 USA
3Chemistry Department, School of Literature, Art and Science, University of Michigan at Ann Arbor,
Ann Arbor, MI 48109 USA
4Center for Biological Nanotechnology, Department of Internal Medicine, University of Michigan Medical School
Ann Arbor, MI 48109-0648 USA
This is an abstract
for a presentation given at the
11th
Foresight Conference on Molecular Nanotechnology
A novel nanostructure was constructed using two different generations of polyamidoamine (PAMAM) dendrimers and complementary 50 base pair oligonucleotides. The oligonucleotides were covalently conjugated to partially acetylated generation 5 and 7 PAMAM dendrimers, and analyzed using UV-vis spectroscopy. These conjugates were then mixed at appropriate temperatures to allow for DNA-directed self-assembly of supramolecular arrays. Overall size and shape of the DNA-linked dendrimer clusters was examined using tapping mode atomic force microscopy (AFM) to resolve the interspatial distance between the linked G7 and G5 dendrimer units. The intra-dendrimer distance was measured to be 21 ± 2 nm, which is in agreement with the theoretical length of the 50-base long oligonucleotide pairs. In contrast, an AFM image of a mixture of non-complexed G7/G5 dendrimers only showed a few dendrimers physically in contact with a inter-dendrimer distance of 8 ~ 10 nm. In addition, a dynamic light scattering (DLS) analysis of the cluster showed a diameter of 37 ± 4 nm, which was the expected diameter of a DNA-linked dendrimer cluster. These results suggest that PAMAM dendrimers can be self-assembled via complementary oligonucleotides to form supramolecular nanoclusters.
KEYWORDS. Oligonucleotide, DNA, PAMAM dendrimer, Self-assembly, Atomic force microscopy.Table of Contents Graphic
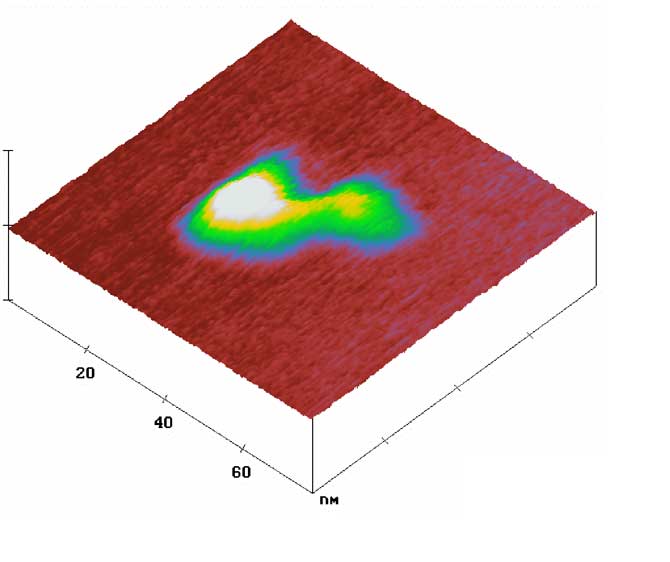
Figure. A representative three dimensional AFM image of a DNA-linked G7-G5 dendrimers on mica (80 nm × 80 nm). The intra-dendrimer distance was measured to be 21 ± 2 nm, which is in good agreement with the theoretical length of the 50-base long oligonucleotide duplex used to connect the polymers. A dynamic light scattering (DLS) measurement of a population of these clusters showed a mean diameter of 37 ± 4 nm.
Abstract in Microsoft Word® format 385,350 bytes
*Corresponding Address:
James R. Baker Jr.
Department of Biomedical Engineering, School of Engineering
University of Michigan at Ann Arbor
9220 MSRB III, 1150 W. Med. Ctr. Dr.
Ann Arbor, MI 48109 USA
Phone: 734-647-2777 Fax: 734-936-2990
Email: [email protected]
|