Molecular Specification via a Simple Scripting Language
by
Peter C. McCluskey
Software consultant
Mountain View, CA 94040
[email protected]
http://www.rahul.net/pcm/
This is a draft paper presented at the
Sixth
Foresight Conference on Molecular Nanotechnology.
This paper is also available on the author's Web site at
http://www.rahul.net/pcm/mmtk/mmtkmnt6.html
Abstract
Chemists have traditionally been mainly concerned with the study of natural molecules, which has led them to give little attention to the means of constructing descriptions of large artificial molecules. Nanotechnology designs will need ways to put together standard components to describe sophisticated systems.
This paper describes software under development that should make it easy to specify complex artificial molecules by simple scripts which describe moieties and how they attach to each other.
Each moiety description includes a list of dangling bonds plus a dihedral angle to specify the orientation of each dangling bond. The relative positions of the atoms in the moiety are specified by the bonds between them and as many dihedral angles as are needed to make it unambiguous.
The scripts are written in Python and use objects from Konrad Hinsen's Molecular Modelling Toolkit, which produces a powerful and flexible language without making the scripts hard to read or reinventing much syntax.
Motivations
When I set out to play with some molecular modelling software, I was
disappointed to find that most of what I found was written around the
"take what you see in nature and analyze it" approach, rather than an
engineering approach to specifying molecules. This convinced me that
if I wanted good software to enable me to create the kind of molecules
I wanted to play with, I would need to write it myself.
Some people who think about nanotech design software focus on visual
ways of specifying molecular systems.
I doubt that a primarily graphical approach will scale up adequately.
Small proteins or Drexler's example parts
appear to be stretching the limits of what I can comprehend via a single
picture. There are severe limits to how accurately I can position objects
using a mouse when trying to control 6 degrees of freedom. GUI's impose
important limitations on parallel operations that we might want to do on
many disparate parts of a system.
The specific problem that provoked me into starting this software was the
desire to find a good molecular equivalent to LegoTM Blocks.
I chose MMTK as the starting point for my efforts because it had the
best combination of general purpose molecular modelling code, readability,
and flexibility. Python's ability to import and parse code at runtime
is an added bonus, which avoids the need to invent yet another scripting
language.
The main drawback of Python is inadequate provision for type checking,
which can lead to confusion and errors in passing objects to imperfectly
documented parts of the system.
Unlike most software tools where there is a clear dividing line between
the user interface and the internal implementation, the use of Python and
MMTK provides a high-level interface designed specifically for most end users,
but also allows sophisticated users access to most low-level
methods in the system.
Users will need to learn some of the Python programming language, but
it is unlikely that they will need to learn more than 5% of what it would
take to become a serious Python programmer.
Overview of the Construction Process
The basic idea behind this software is that one should build up a modest
library of simple moieties, and then describe how these moieties are bonded
together or positioned relative to each other to form large systems.
Creating a Moiety
Create a file in the Groups subdirectory of your MMTK installation.
Start by creating the atoms using statements such as:
C1 = Atom('C')
(C1 is a name that will be used later to refer to this carbon atom later).
Then create bonds that connect the atoms using statements such as:
b12 = Bond(C1, C2)
This will sometimes be enough to specify the moiety shape, but more often you
will need to constrain some of the dihedral angles (also known as torsion
angles - which describe the angles between the plane of the first 3 of 4
atoms and the plane of the last 3 atoms) using statements such as:
da1 = Dihedral(C1, C2, C3, C4, 180*deg)
For moieties that are likely to be used with the Amber force field as it
is currently implemented in MMTK, you will need to specify a mapping of
atom names to Amber types such as:
amber_atom_type = {C1: 'CT', C2: 'CT', C3: 'CT', C4: 'CT' }
I have implemented an MM3 force field which can infer the correct atom
types in many circumstances, and I hope that in the future the need for
explicit atom type lists will become increasingly rare.
Next you need to list places where bonds can be attached to the moiety.
The PrintDanglingBonds.py utility program will produce a list of statements
such as:
dC3a = DanglingBond(C3, Dihedral(C1, C2, C3, None, -60.329439*deg))
which may be good enough, although it's probably a good idea to round
the angles it produces to nicer numbers such as -60. The None in
the Dihedral indicates where an atom from another moiety can be attached.
Finally, you should group the DanglingBond's into ordered sets
with descriptive names to make it easier to specify how to attach
other moieties:
hookwest = Hook(None, [dC1b, dC3a])
hooknorth = Hook(None, [dC1c])
(The None's in these examples don't serve any real purpose, and
will probably become unneeded in a future version.
A simple example of a script to attach moieties together:
block1 = Group('diamondoid_fragment') # read in moiety
join = Joiner(block1) # create object that does joins
join.repeatGroup('hookeast', 'hookwest', 5) # create 5 copies of block1
# joined by hookeast, hookeast
result = join.getResult() # tell Joiner it's finished
result.setPositions(forcefield) # assign coordinates to atoms
result.view() # display results
To attach two different types of moieties together instead of repeating one
type, you would use a joinGroups method call instead of repeatGroup:
join.joinGroups(block1.hookwest, moiety2.hook1)
Detailed Example
This example shows how my search for a molecular Lego (tm) block has
been proceeding.
I start by creating the following straightish 4 carbon moiety:
name = 'diamond4carbon'
C1 = Atom('C')
C2 = Atom('C')
C3 = Atom('C')
C4 = Atom('C')
b12 = Bond(C1,C2)
b23 = Bond(C2,C3)
b34 = Bond(C3,C4)
da1 = Dihedral(C1,C2,C3,C4,180*deg)
amber_atom_type = {C1: 'CT', C2: 'CT', C3: 'CT', C4: 'CT' }
dC1a = DanglingBond(C1, Dihedral(None,C1,C2,C3,60*deg))
dC1b = DanglingBond(C1, Dihedral(None,C1,C2,C3,-60*deg))
dC1c = DanglingBond(C1, Dihedral(C3,C2,C1,None,180*deg))
dC2a = DanglingBond(C2, Dihedral(None,C2,C3,C4,120*deg))
dC2b = DanglingBond(C2, Dihedral(None,C2,C3,C4,-120*deg))
dC3a = DanglingBond(C3, Dihedral(C1,C2,C3,None,60*deg))
dC3b = DanglingBond(C3, Dihedral(None,C3,C2,C1,120*deg))
dC4a = DanglingBond(C4, Dihedral(C2,C3,C4,None,60*deg))
dC4b = DanglingBond(C4, Dihedral(C2,C3,C4,None,-60*deg))
dC4c = DanglingBond(C4, Dihedral(C2,C3,C4,None,180*deg))
hooknorth = Hook(None, [dC1c])
hooksouth = Hook(None, [dC4c])
hookup = Hook(None, [dC2b, dC4b])
hookdown = Hook(None, [dC1a, dC3b])
hookeast = Hook(None, [dC2a, dC4a])
hookwest = Hook(None, [dC1b, dC3a])
Most of the remaining code in this example comes from a script named
lego6.py, and
roughly corresponds to the chronological steps by which I proceeded.
Each excerpt is followed by some explanations.
ff = GenericForceField('mm3')
world = InfiniteUniverse(ff)
block1 = Group('diamondoid_fragment')
join = Joiner(block1)
width = 5
join.repeatGroup('hookeast', 'hookwest', width)
join.repeatGroup('hooksouth', 'hooknorth', 5, 1, [180*Units.deg]*width)
join.repeatGroup('hookup', 'hookdown', 2)
result = join.getResult()
result.setPositions(ff)
The name result is now bound to a Group that is a roughly
rectangular slab of 5 by (4*5)=20 by 2 carbons. The 1 in the
middle repeatGroup call is an optional argument that controls
whether the repeated block is reused or cloned. The list of width
angles after it specifies the angles for the dihedrals about the single bond
that connects each hooksouth Hook to each hooknorth.
Ideally the software should be able to deduce that those are the only
reasonable dihedral angles for those bonds, but it will take more programming
effort to find a way of doing this that scales up well.
The other hooks in this example all involve more than one bond, which
automatically constrains the dihedral in a way that the software can
deduce if the constraints are consistent with each other.
a_hole = result.findAtomByName('diamond4carbon.diamond4carbon7.C4')
surface_point = Positioner.findNearestPointOnSurface(result, a_hole)
surface_vector = surface_point - a_hole.position()
dist_to_surface = surface_vector.length()
surface_dir = surface_vector.normal()
cylinder1 = Cylinder(a_hole.position() - 0.1*Units.Ang*surface_dir,\
2.5*Units.Ang, (1*Units.Ang+dist_to_surface)*surface_dir)
I want to dig a hole in one face of the slab. I looked at what the script
had produced so far (using a version of RasMol that I
have begun to integrate into MMTK), decided on an atom to center the hole
around, and got it's name by clicking on it. I then created a line from
that atom through the nearest point on the convex hull of the system (a set
of triangles bounding the atom positions as closely as possible without
making any concavities), and generated a cylinder about that line (extending
the line a bit to make sure it included that atom and any other that might
be on the cylinder boundary.
atoms_in_cyl1 = result.atomsWithinRegion(cylinder1)
result.removeAtoms(atoms_in_cyl1, None)
This removes the atoms enclosed by the cylinder.
c_below = result.findAtomByName('diamondoid_fragment.diamondoid_fragment32.C3')
nitrogen = result.generateNewAtom('N')
result.replaceAtoms([c_below], [nitrogen])
result.fillDanglingBonds(ff, 'N', 8, 3)
I saw that the carbon atom remaining at the center of the hole's bottom had a
dangling bond remaining that might interfere with the way I hoped to insert
things into the hole, so I replaced it with a nitrogen.
Six places where carbons had been removed left places to which 3 dangling
bonds were pointing; the fillDanglingBonds call finds all such
positions and fills them with atoms of symbol 'N', MM3 type 8.
def filter2n(atom): # define a function which returns true
if atom.symbol != 'C': # for carbons bonded to exactly 2 nitrogens
return 0
bonded_to = atom.bondedTo()
countn = 0
for a in bonded_to:
if a.symbol == 'N':
countn = countn + 1
return countn == 2
carbons_bonded_to_2_ns = filter(filter2n , result.atomList())
selected_ns = []
replace_cs = []
for a1 in carbons_bonded_to_2_ns:
for a2 in a1.bondedTo():
if a2.symbol == 'C':
c = a2
a1_pos = a1.position()
for a2 in a1.bondedTo():
if a2.symbol == 'N':
x_product = (a1_pos - c.position()).cross(a2.position() - a1_pos)
if surface_dir * x_product > 0: # select by orientation
selected_ns.append(a2)
replace_cs.append(result.generateNewAtom('C'))
result.replaceAtoms(selected_ns, replace_cs, ff)
for i in range(len(carbons_bonded_to_2_ns)): # make double bonds:
result.attachBondBetween(carbons_bonded_to_2_ns[i], replace_cs[i])
I want to create some double bonds along the edge of the hole to make
it moderately stable by itself, but still able (I hope) to react with
similar double bonds on the knob I plan to insert into the hole.
First I search for all the carbon atoms attached to two nitrogens, which
gives me three carbons associated with the six nitrogens I added in the
prior step. I then identify three of the nitrogens on the basis of an
arbitrarily chosen orientation from the selected carbons, replace those
three nitrogens with carbons, and create double bonds from the previously
selected carbons to the new carbons. The double bonds will be somewhat
strained, but probably no more so than in figures 8.9 and 8.10 in
NanoSystems (cubene and adamantene).

I'm now satisfied with the hole, and turn my attention to adding a knob
on the other side of the slab that will fit into such a hole on another
"Lego block". I create the following file cyanide:
name = 'Cyanide'
C = Atom('C')
N = Atom('N')
b = Bond(C, N, 3)
mm3_atom_type = { C : 4, N : 10 }
db = DanglingBond(C, Dihedral(None, N, C, None, 0.0))
hook1 = Hook(None, [db])
Back to the main script:
hook3_list = []
for db in result.danglingbonds:
if nitrogen in db.a1.bondedTo():
hook3_list.append(db)
hook3 = Hook((result, hook3_list))
join2 = Joiner(result)
layer = Group('lego4')
join2.joinGroups(hook3, layer.hook1, 0)
result = join2.getResult(0)
This creates a knob of three nearby cyanides. (Possibly the nitrogens will
bond with neighboring cyanide carbons to form a ring. That would probably be
equally close to the shape and reactivity I want).
result.setPositions(ff)
result.hydrogenateDanglingBonds(ff)
This defines positions for the atoms in the knob, and adds fills all remaining
dangling bonds with hydrogens.
result2 = result.clone()
result2.name = 'd42'
face2 = []
for a in result2.atomList():
if a.symbol == 'N':
bonded = a.bondedTo()
if len(bonded) == 1 and bonded[0].symbol == 'C':
face2.append(a)
face2 = [face2[0], face2[2], face2[1]]
PositionAdjacent(result,carbons_bonded_to_2_ns, result2,face2, [2*Units.Ang]*3)
Finally, I duplicate the molecule, pick three atoms on the knob of the
new molecule, and rotate/translate it so those three atoms are 0.2 nanometers
(2 angstroms) from three of the carbons on the edge of the original molecule's
hole (actually, it can't satisfy those three constraints, and the actual
distances are 0.2 to 0.32 nanometers (2 to 3.2 angstroms); see the
PositionAdjacent description
for details).
This produces a pair of molecules with potentially reactive double
and triple bonds positioned 0.3 to 0.4 nanometers (3 to 4 angstroms) apart.
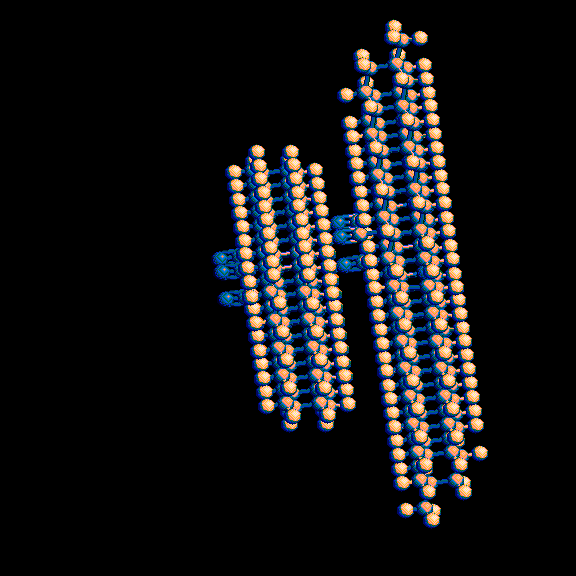

3-D representations of these images are available at
http://www.rahul.net/pcm/mmtk/mnt6_images.html.
I plan to add another hole and another knob at the other end of the slab
and run molecular dynamics simulations on the result to determine whether the
three double bonds / triple bond pairs react to bond the two molecules together
in a reasonably rigid result, and if so how closely they have to be positioned
before they spontaneously form these bonds in a predictable one of the three
possible orientations (each 120 degrees apart). If that works, I will tweak
the shapes a bit so they fit well when I put many of them together into
larger structures.
Prospects
If such building blocks could be created before a real assembler exists
(a big assumption which I know I can't justify), then a positioning device
with a fair amount of error might be able to construct a wide variety of
atomically precise rigid structures.
I haven't yet addressed constructing systems which are capable of motion.
I started with diamondoid systems because they are simpler to model, but
I am equally interested in solution-based systems.
Will this approach work for floppy molecules such as DNA or proteins?
I expect some problems associated with handling under-constrained dihedrals.
I don't want to tackle the protein folding problem, but I also want to
minimize the need for specifying dihedrals by users who are constructing
systems from standardized components.
When I have the diamondoid tests sufficiently debugged, I will try designing
some building blocks using DNA that have some hope of producing controlled
motion and of being produced with today's technology.
Future Software Enhancements
- Easier ways of specifying moieties
While I believe my initial approach of relying on dihedrals to specify
shapes provides a powerful method of indicating which parts of the
moiety need to be constrained without constraining things which should
adapt to the conditions under which it is being used, it has proven harder
that I expected to use them to design good components.
At very least I will need to supplement this with the ability to import
PDB files as rigid components. I will also try to find reasonable ways
to relax the rigidity requirement without allowing the software to guess
at absurd conformations.
I may try include using distances between atoms to specify constraints.
- Search and replace
The existing replaceAtoms function, combined with simple pieces of Python
code, provides some of what I want in a search and replace function, but
only works on single atoms so far, and the search part is an accidental
byproduct of the power created by MMTK's approach. I plan to investigate
whether I can produce a regular expression language that improves on this
search approach.
- Enhanced Geometric Specifications
I plan to support using a wide variety of geometric objects, including
unions and intersections of geometric objects (constructive solid geometry
as used in CAD software), plus the ability to fill regions defined by
molecular shapes read from PDB files with user-defined patterns of atoms.
- Speed
Currently the joining and positioning steps in the example above
take about 150 seconds on a 300 mHz Pentium II, and appear to scale up
roughly in proportion to the square of the number of atoms. I plan to
identify the bottlenecks and replace them with O(n) or O(n log n) algorithms
where possible.
- Modes that are more Interactive
Currently I use the system as a compiler, rebuilding most moieties
from scratch each time I test something. It should be possible to use
it more like an interpreter (saving intermediate steps) with many objects
an operations on them selected via a GUI (presumably derived from RasMol)
whenever that can be made quicker than typing the text-based methods.
- Accumulate a library of standard moiety components.
- Absolute coordinates
I have been trying to minimize use of absolute reference frame in favor of
relative positions, partly in hopes of being close to how an actual assembler
would operate, but mainly because the setPositions method includes
some random decisions which ensure that no 2 runs produce the same atom
positions. I will try to find out why seeding the random number generator
doesn't make the arbitrary choices repeatable, as that will probably make
some geometric operations easier.
- More work is needed on error messages. Inconsistent dihedral specifications
for a ring structure tend to produce low-level complaints such as:
Warning: Nearest available angle 48.24 degrees from expected
Dihedral(Atom Lego3.C1,Atom Lego3.N1,Atom
Lego3.C2,None,-60.0*deg); expected -60.00
Traceback (innermost last):
File "Programs/lego1.py", line 32, in ?
result.positionDanglingBonds(ff)
File "./CompositeObject.py", line 1772, in positionDanglingBonds
(dir, constraint, cmbond, far_bond) =\
File "./CompositeObject.py", line 985, in nextBondDirection
raise ConformationError
ConformationError
where what is really needed is a way to identify what constraint was
inconsistent with the constraint it failed to satisfy here.
Reference Manual
There is no well-defined line between what methods are intended for users
writing scripts that construct systems, and methods that are intended for
programmers implementing more methods, but this section will try to give
specifications of those methods that are most useful for script writers.
Also, the toolkit is still in it's infancy as this paper is being written;
this version is designed more to provide a taste of what the system can
accomplish than as a programming guide.
Updated information will be available at
http://www.rahul.net/pcm/mmtk/,
along with more example scripts and pdb files they produce
(such as "Hello, world!" written in hydroxyls on diamondoid),
and source code.
Classes/Methods
Notations used:
- Anything not listed under a class is a subroutine. Anything listed
under a class is a method of that class (a constructor if it's name is
the same a the class name) or a data member (if no parentheses)
- Classes listed in parentheses after a class X are parent
classes of that class.
- Optional arguments are listed with " = ", default values.
- All distances are in nanometers.
- All angles are in radians.
References
- Drexler, K.E. (1992) NanoSystems, Wiley-Interscience. Sections 8.4 through 9.5.
- Leach, A.R. (1996) Molecular Modelling, Addison Wesley Longman.
- Preparata, F.P.; and Shamos, M.I. (1985) Computational Geometry,
Springer-Verlag.
Describes optimal speed geometric algorithms (oriented more towards theoretical
computer scientists than towards laymen).
Lego is a trademark of the LEGO Group.
Corresponding Address:
Peter McCluskey
1571 El Camino Real W.,#15
Mountain View, CA 94040
(650) 967-3359
[email protected]
http://www.rahul.net/pcm
|