Axonemal dynein -- a natural molecular motor
by
Helen C. Taylor* and Michael E.J. Holwill
Physics Department, King's College London, Strand, London WC2R 2LS, UK
*Corresponding author: [email protected]
This is a draft paper for the
Sixth
Foresight Conference on Molecular Nanotechnology.
The final version has been submitted
for publication in the special Conference issue of Nanotechnology.
Summary
Biological motor molecules in vivo possess many of the characteristics required to power nanomachines. They can generate force and torque, transport specific cargoes over appropriate substrates, and the character and rate of their action can be controlled. In cilia and flagella, axonemal dynein motor molecules are attached to nine microtubule doublets arranged cylindrically around a pair of single microtubules. The dynein motors undergo a cycle of activity, during which they form a transient attachment to the doublet, and push it towards the tip of the cilium or flagellum. The microscopy techniques currently available do not have sufficient resolving power to view this activity directly. Instead, movement of microtubules by the action of assemblies of isolated dynein arms activated by ATP can be studied in vitro. At a particular ATP concentration, microtubule gliding velocities are found to increase with microtubule length. By making appropriate assumptions about the system, it is possible to predict its behaviour using computer simulations. These simulations allow us to investigate certain properties of individual dynein molecules in addition to characterising the co-ordination of activity within the assembly of arms. Agreement between the experimental results and computer predictions can be achieved by selecting appropriate characteristics of individual dynein arm action for either random or systematic activity of the arm assembly. Based on our computer simulations, it is possible to design experiments to differentiate between the co-ordination patterns. We have also simulated microtubule sliding for comparison with the sliding which occurs when microtubules are extruded from disintegrating cilia and flagella. These studies lay the foundation for the development of computer models of the whole axoneme, which will allow us to investigate the way in which the dynein motors interact with other structures to produce ciliary and flagellar bending.
Introduction
There are three families of naturally-occurring motor molecules: the myosins, the kinesins and the dyneins. All function by undergoing shape changes, utilising energy from the biological fuel adenosine triphosphate (ATP). Each family has members that transport vesicles through the cell cytoplasm along linear assemblies of molecules -- actin in the case of the myosins and tubulin for both of the other families. The kinesins and dyneins move or 'walk' along microtubules - tubes constructed from tubulin - carrying their cargo. The microtubules are polar structures -- only one end (the plus end) is capable of rapid growth by adding more tubulin molecules -- and the action of the motor molecules is polarised so that they move in one direction only. Kinesins are plus-end directed motors whilst dyneins move towards the minus end of the microtubule, which tends to be anchored in the centrosome of the cell. Motor molecules are also found in the contractile elements (myofibrils) of muscle cells, and in cilia and flagella, the oscillatory, whip-like appendages of a range of cells. The motive power for muscle activity is provided by myosin motors, organised as thick filaments which interact with an array of thin actin filaments to cause the shortening of elements within each myofibril. This shortening is achieved by relative sliding of the myosin and actin filaments. Dynein motors also cause sliding between microtubules that form the skeleton of cilia and flagella. Other ciliary structures resist this sliding with the result that bends form along the length of the cilium or flagellum and propagate from base to tip (e.g. mamalian sperm flagella) or from tip to base (e.g. flagella of Crithidia oncopelti, a parasite of the milk weed bug). The propagation of these bends requires co-ordinated action of the several types of dynein motor present in the cilium or flagellum and the structures providing the resistance to sliding. In this paper we will review our investigations into how these dynein motors operate in the cilium or flagellum, which itself is a micromachine (typically 5 m m long) capable of propelling cells.
Location of axonemal dynein
Cilia and flagella are found in organisms throughout the plant and animal kingdoms; specific examples are cilia in the human lung, where they move mucous, and the flagella used to propel mammalian spermatozoa. The axoneme (Fig. 1) is the core structure of a cilium or flagellum, and consists of a set of nine doublet microtubules -- each of which consists of a partial microtubule connected to a complete microtubule -- arranged cylindrically around a pair of singlet microtubules. These microtubules are arranged with their plus ends at the tip of the axoneme and their minus ends anchored in basal bodies in the cell. The axonemal dynein motors are distributed along each doublet as inner and outer rows of arms (Fig. 1).
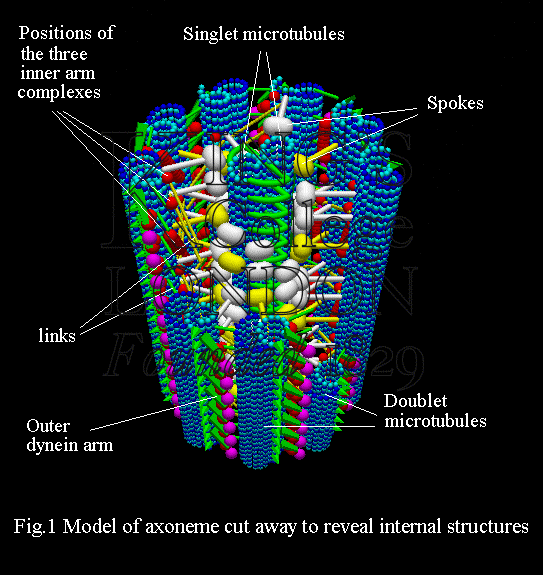
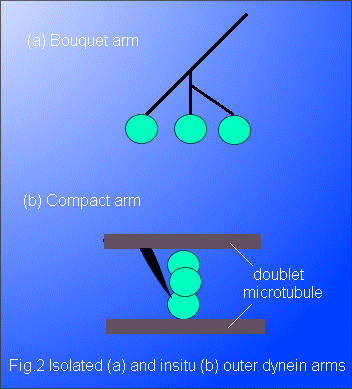
When removed from the cilium or flagellum, and examined by electron microscopy, the outer dynein arm appears as a bouquet structure consisting of a branched stalk bearing, in general, three globular heads (Fig. 2a). Each head is about 10 nm in diameter and is formed from a long, folded molecular chain with a weight in the range 450 to 550 kDa; the stalk is made up of light and intermediate molecular weight chains. Electron microscopy has revealed that, within the axoneme, the outer dynein arm adopts a more compact configuration (Fig. 2b) than the bouquet, with the stalk forming a cape which has one end permanently attached to a doublet. The electron micrographs obtained show several distinct conformations of the outer arms. Many of the micrographs (Avolio et al. 1986; Sugrue et al. 1991) support the interpretation that the heads are mounted on each other so that the arm is able to bridge the interdoublet gap of about 24 nm, thereby allowing one head to interact with the neighbouring doublet. The inner arms, with more than seven chemically-different heavy chains arranged in three distinct complexes, have a more complicated structure than the outer arms and will not be discussed here in detail.
Behaviour of outer dynein arms in vitro
The observation techniques currently available do not have sufficient resolving power to allow an individual dynein arm to be seen in action. However, the behaviour of assemblies of these arms can be inferred through their effects on larger, resolvable structures. We use computer-modelling techniques to interpret these experimental observations in terms of the co-ordination of arms within the assemblies, and hence to gain information about the characteristics of a single arm.
Using photographic and video-enhanced microscopy techniques, the movement of microtubules resulting from dynein arm action has been studied. In the following sections several experiments will be described and interpretations of the results based on our computer models discussed.
Gliding microtubule studies
Outer dynein arms can be extracted from the axonemes of some cilia and distributed randomly over a glass surface with a density that can be determined experimentally. Hamasaki et al. (1995) have shown that when microtubules are dropped onto the motors and ATP added, the microtubules are transported over the surface with velocities that depend on microtubule length (Fig. 3). All microtubules glide in a direction parallel to their longitudinal axis and in experiments where the polarity of the microtubules is known (Yamada et al. 1998), they glide minus-end (or tip) first. These observations imply that, in the vicinity of the microtubule, each dynein arm is attached to the glass surface and may undergo a cycle of activity during which it forms a transient attachment to the microtubule and pushes it tipwards. The unidirectional character of microtubule gliding suggests that motor action is polarised in such a way that the force is applied in one direction along the microtubule axis.
Using an approach based on the Briggs-Haldane theory of chemical kinetics, a hyperbola (equation 1) was fitted to the velocity (V) versus microtubule length (L) data (Fig. 3) obtained from the in vitro motility assay (Hamasaki et al. 1995).
............ (1)
In this equation, V0 is the maximum velocity and KL is a constant equal to the microtubule length at which the velocity of microtubule translocation is half its maximum value. The period of attachment of a motor to its molecular 'track' is known as the duty phase, and can be estimated using equation (2), which was derived by Uyeda et al. (1990) for actin filaments gliding over a field of myosin molecules. In this equation, f is the fraction of the cycle time for which the motor is attached to the gliding filament, while N is the number of motors available to interact with the filament.
............ (2)
After calculating the number of arms which could interact with a microtubule of length KL, this equation was used to predict the fraction, f, of the cycle occupied by the duty phase of dynein. For this in vitro motility assay f was found to be 0.01. It should be noted that the scatter in the experimental data (Fig. 3) leads to uncertainty in the values of the parameters (including KL) used to describe the fitted curve, and hence to uncertainty in f.
The collective and individual behaviour of the motors along the length of the gliding microtubules has been examined in our laboratory using computer simulation techniques. Since a random distribution of dynein arms characterises the experiment, it is likely that each arm capable of interacting with the microtubule is randomly activated in time. In the gliding experiments, the average separation of dynein arms on the substrate was found to be 60 nm. To simulate the activity of the assembly of arms, appropriate assumptions about the gliding microtubule system were made. One assumption is that the system behaves as if it were unloaded; this is reasonable since the viscous resistance experienced by the moving microtubule is significantly less than the driving force provided by the dynein arms. This assumption implies that the microtubule glides with the velocity of the driving dynein arm during its attachment phase. The distance that the arm can push the microtubule during the attachment phase is the step size, and has yet to be thoroughly examined experimentally; for our simulations it was taken to be 16 nm. A number of simulations of the random activity of dynein arms was performed to predict gliding velocities at various microtubule lengths. In each simulation the attachment phase of the dynein arm occupies 1% of the complete cycle (i.e. f = 0.01). The gliding velocity is assessed over a single cycle time at which point every dynein arm in the field of 1000 arms will have completed a cycle. The phase at which each arm begins its cycle is allocated randomly using a standard random number generator. A graph of velocity of microtubule translocation as a function of microtubule length is shown for random arm activation in Fig. 4. As the length of the microtubule increases the chance of the microtubule interacting with an arm becomes greater, leading to a rise in the simulated gliding velocity. The graph reaches a plateau when the microtubule is of such a length that it could potentially be in contact with at least one active arm throughout the simulation. In these simulations we have assumed that when two or more arms interact simultaneously with a microtubule, it glides through a distance of only one step size. This accounts for the maximum gliding velocity observed. The scatter of the simulated data is due to the random activation of each dynein in the assembly of motors.
The experimental and simulated data presented respectively in Figs 3 & 4 show similar trends, with the velocity of sliding increasing asymptotically with microtubule length to a maximum. There is considerable scatter of the experimental data in Fig. 3, which could be the result of physical effects of the motors or may arise from experimental error. The simulated data set (Fig. 4) shows scatter arising from the random activation of motors in the assembly, but it can be observed that the character of this scatter differs from that in Fig. 3. A quantitative examination of the two data sets reveals that the gliding velocities predicted by the simulations are an order of magnitude greater than the experimental values. It will therefore be necessary to investigate the effects of uncertainties in the parameters f and step size on the predictions of the simulation.
When the microtubules are dropped onto a field of dynein molecules in the absence of ATP, the microtubules are observed to remain stationary rather than to drift with Brownian motion. This observation suggests that there is weak attraction between the microtubule and the nearby dynein. The addition of ATP activates the dynein and the microtubule is seen to glide in the direction of its longitudinal axis. These observations suggest that co-ordination may be imposed by the microtubule itself. In such a co-ordination mechanism, each dynein arm could be triggered into action by a signal, carried by the microtubule, from its immediate neighbour, so that the arms are activated in a sequential manner. In this case a wave of activity sweeps along the doublet. A computer model of this co-ordination was developed in our laboratory based on the assumptions listed above for the random model. Initially the relationship between gliding velocity and microtubule length was investigated for a step size of 16 nm and f = 0.01, and the results are shown in Fig. 5. The average velocity of the microtubule increases with microtubule length until this length is such that the microtubule is always in contact with at least one active dynein arm; this length corresponds to one complete wave of dynein arm activation. At this and greater lengths the velocity remains constant. In the computer simulation, the transition from an increasing to a constant velocity occurs abruptly (Fig. 5). This abrupt transition is not apparent on the experimental plot in Fig. 3 and the scatter seen in this data set is not predicted by the model. Detailed comparison of the experimental and simulated data reveals an order of magnitude difference between the two maximum velocities. As discussed in an earlier section, values for several parameters, including the step size and f, used in the simulation are subject to some uncertainty. Comparison with kinesin, the other microtubule motor, suggests that the step size for dynein could be as short as 8 nm. Using this value in the simulation results in a very similar relationship (Fig. 5) between the microtubule velocity and length to that for a step size of 16 nm (with one complete wave of dynein arm action occupying the same length). The magnitudes of the velocities are reduced, but are still an order of magnitude larger than those obtained experimentally. f, the second parameter subject to uncertainty, was increased by a factor of 10 and the simulation repeated at both step sizes. The graphs in Fig. 5 indicate that under these conditions the gliding velocities are reduced to the same order of magnitude as those observed experimentally. The simulation at a step size of 8 nm with f = 0.1 produced velocities comparable to the experimental values although, as discussed earlier, some features of the experimental distribution (e.g. the scatter) are not reproduced by the simulation.
Sliding microtubule studies
Suitable preparative chemical treatment, which presumably removes the linking structures shown in Fig. 1, allows individual axonemes to disintegrate on addition of ATP. Examination of the preparation by both light and electron microscopy reveals that the doublet microtubules have slid relative to one another out of their cylindrical array. This sliding disintegration can also be initiated in flagella from mutants of Chlamydomonas (a single-celled organism found in ponds) from which the radial spokes and the complex associated with the central pair of microtubules (Fig. 1) are missing. This supports the hypothesis that dynein interaction between the doublets is responsible for the sliding.
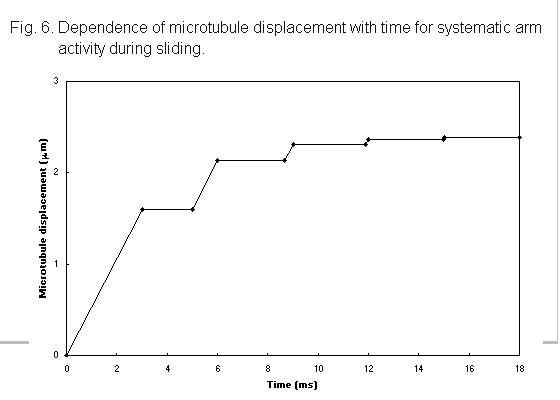
Observations reveal that sliding occurs in one direction only, indicating that the polarisation observed for isolated arms is retained when the arms are attached to microtubules (Sale & Satir, 1977). Quantitative experimental analyses of sliding are at present incomplete. Takahashi et al. (1982) published observations of the distance moved by one microtubule relative to another during sliding, over time periods of up to 4 seconds; both outer and inner dynein arms were present in these disintegrating axonemes. The results show that the microtubule displacement increased linearly with time, although deviations from the linear relationship were recorded within either the initial or the final second of the motion.
In these sliding experiments, the movement of a microtubule is produced by the action of an ordered array of dynein molecules, separated by 24 nm in the case of the outer arms, on the neighbouring doublet (Fig. 1), rather than by a random arrangement as in the gliding experiments. Because of the structured organisation, it is not unreasonable to suggest that the arm action is highly co-ordinated, rather than stochastic as is probable in the gliding experiments. One of the possible co-ordination mechanisms for the outer arms is that described in the previous section, where sequential waves of dynein activity sweep along the doublet. As sliding progresses, the length of overlap, i.e. the region of dynein arm interaction, between the microtubules decreases. While this region is longer than the wavelength of activation, there will always be at least one dynein arm interacting with the driven microtubule, which will therefore move with a constant velocity, as is seen in the experimental plots of Takahashi et al. (1982). When the length of the overlap falls below that of the wavelength, there will be periods of time when no arms are interacting and the driven microtubule will be stationary. The driven microtubule will then move in a stepwise manner, as shown in Fig. 6. With a dynein arm separation of 24 nm and an f value of 0.01, the activation wavelength is 2.4 mm; if this, or a similar value, holds in the experimental arrangement, observation techniques have sufficient resolution that the nature of the microtubule motion for an overlap region of this size could be determined. One of the curves published by Takahashi et al. (1982) shows some evidence of a stepwise motion, but further critical investigations of microtubule movement for short overlap regions is needed before definite conclusions can be drawn.
Discussion
The random and co-ordinated mechanisms of outer arm activity in the gliding experiment, examined using our computer techniques, have generated velocity versus microtubule length relationships which show trends similar to those of the experimental data, but they do not predict the scatter satisfactorily (Fig. 3). Both mechanisms show quite different behaviour as the maximum velocity is approached, with a smooth rise to an asymptotic value characterising the stochastic activity (Fig. 4), while, when the action is co-ordinated, there is an abrupt change to a constant velocity at a critical length equal to the wavelength of dynein arm activation (Fig. 5). The scatter of the experimental data is such that differentiation between these activity mechanisms is not possible. Further experimental data are required to confirm that the scatter is a product of dynein action rather than of experimental error. One characteristic of the dynein substrate that may account for this scatter is associated with the random orientation of the dynein arms. Although their action is polarised such that the resultant force generation, and hence displacement, is towards the minus end of the microtubule, the random orientation of the arms may cause the force to be applied at various angles to the longitudinal axis of the doublet. We believe that the variety of displacements produced in this way may be responsible for the experimental scatter, and this is currently being studied. The consequence of uncertainty in both the step size and f was investigated in the simulations of gliding induced by co-ordinated arm activity. Increasing the duty phase and decreasing the step size reduced the value of microtubule velocity to a magnitude comparable with the experimental data. We predict that implementing such changes in the random activity simulations will also reduce the velocities predicted whilst maintaining the form of the relationship between velocity and microtubule length. Kinesin, the other microtubule motor, has a step size of 8 nm (Svoboda et al. 1993), so this value is not unreasonable for dynein, and a recent study (Shingyoji et al. 1998) suggests that the duty phase may be longer than previously thought.
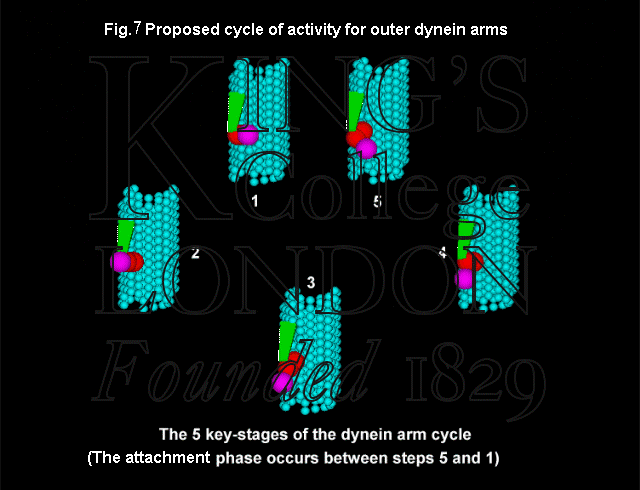
The sliding simulations represent the activity of the outer arms only, whereas the limited experimental observations (Takahashi et al. 1982) described earlier involve axonemes disintegrating under the action of both inner and outer arms. While the structure and activity of the outer arms has been reasonably well established on the basis of electron microscopy (Fig. 7), it is only recently that a unifying description of the complex arrangement of the inner arms has been proposed (Taylor et al. 1997). We have proposed cycles of activity for the inner arms, and anticipate that the techniques being developed for the study of inner arm behaviour will allow us to refine our computer models of the system. For example, studies of the activity of the seven inner dynein heavy chain molecules using microtubule gliding techniques have shown that many of the molecules are capable of translating and, in some cases, rotating microtubules (Kagami & Kamiya, 1992).
For ease of comparison with the predictions of computer simulations, it would be valuable to have observations of disintegrating mutant Chlamydomonas flagella with rows of inner or outer motors missing; motile characteristics of such mutants have been described by Brokaw & Kamiya (1987). Our computer predictions also suggest that an important investigation is a critical experimental study of the patterns of sliding disintegration when the region of overlap between neighbouring microtubules is small. This type of study is likely to pose technical difficulties as the extruded microtubule experiences forces due to Brownian motion and the influence of neighbouring surfaces.
Implications for in vivo arm activity
The bends propagating along an intact cilium or flagellum reflect the activities of the arms in both rows, requiring a system which controls both the motor activity within the individual rows and the co-ordination between the rows. The in vitro co-ordination studies described above provide data of relevance to the in vivo situation. In the intact cilium, the doublets are tethered at the base, and doublet sliding will result in bend formation. By extending and developing the sliding experiments it should be possible to examine the bends formed in such a situation. Using appropriate computer-modelling techniques we plan to predict the form of bending produced by both random and co-ordinated arm activity. It may be of significance to note that in a typical 100° flagellar bend, the maximum amount of sliding between neighbouring doublets is about 100 nm. With a step size of 8 nm, the activity of about 12 dynein arms would be sufficient to generate the sliding. Within the 100°
bend, which will typically have a length of 5 µm, there will be about 200 pairs of dynein arms on an individual doublet, considerably in excess of the number required to produce the geometrical displacement. While it is also necessary to consider the energy requirements of the system, the potential involvement of only a small fraction of the dynein arm assembly could support the idea that arm activity in the intact axoneme is random.
Our studies should allow us to establish the type of motor co-ordination that exists in the intact cilium, and will also provide information about the behaviour and properties of the interdoublet links (Fig. 1). We plan to use our modelling procedures, in tandem with appropriate experiments, to investigate these control mechanisms with a view to constructing a functional computer model of the complete axoneme.
This paper has demonstrated the sophisticated mechanical properties of just one of the natural molecular motors. Associated research is also in progress to study the chemical attributes of the activity cycles of many of these motors. A thorough understanding of the mechanochemistry of the natural molecular motors clearly has an important bearing on the aspirations of those involved in the design and construction of artificial nanomachines.
Acknowledgements:
Experimental data for Fig. 3 kindly supplied by T. Hamasaki and P. Satir, Albert Einstein College of Medicine. This work was supported by Grant number 29/E09266 from the Biotechnology and Biological Sciences Research Council of the UK.
References
Avolio, J., Glazzard, A.N., Holwill, M.E.J. & Satir, P. (1986) Proc. Natl Acad. Sci. 83:4804-4808.
Brokaw, C.J. & Kamiya, R. (1987) Cell Motil. Cytoskel. 8:68-75.
Hamasaki, T., Holwill, M.E.J., Barkalow, K. & Satir, P. (1995) Biophys. J. 69:2569-2579.
Kagami, O. & Kamiya, R. (1992) J. Cell Sci. 103:653-664.
Sale, W.S. & Satir, P. (1977) Proc. Natl Acad. Sci. 74:2045-2052.
Shingyoji, C., Higuchi, H., Yoshimura, M., Katayama, E., & Yanagida, T. (1998) Nature 393:711-714.
Sugrue, P., Avolio, J., Satir, P. & Holwill, M.E.J. (1991) J. Cell Sci. 98:5-16.
Svoboda, K., Schmidt, C.F., Schnapp, B.J. & Block, S.M. (1993) Nature 365:721-727.
Takahashi, K., Shingyoji, C. & Kamimura, S. (1982) Symp. Soc. Exp. Biol. 35:159-177.
Taylor. H.C., Holwill, M.E.J. & Satir, P. (1997) Mol. Biol. Cell 8:Suppl.51a.
Uyeda,T.Q.P., Kron, S.J. & Spudich, J.A. (1990) J. Mol. Biol. 214:699-710.
Yamada, A., Yamaga, T., Sakakidara, H., Nakayana, H. & Oiwa, K. (1998) J. Cell Sci. 111:93-98.
|