Currently structural DNA nanotechnology, either scaffolded DNA origami or DNA bricks, is the most promising method to build arbitrarily complex multi-million-atom atomically precise structures (see, for example these posts from the past six months Re-engineering a junction to give a new twist to DNA nanotechnology, Testing and improving scaffolded DNA origami for molecular nanotechnology, and Arbitrarily complex 3D DNA nanostructures built from DNA bricks). One of the major limitations of these methods, however, is the cost and quality of the small, single-stranded DNA molecules required, which are prepared by solid state chemical synthesis. Without these solid state synthesis methods, DNA nanotechnology, and indeed much of molecular biology, would have been impossible. However, chemical synthesis is expensive, rendering scale-up very difficult, and as the oligonucleotides become longer than a few tens of nucleotides, impurities and mistakes in synthesis become difficult to remove, potentially compromising atomic precision. Fortunately two DNA nanotechnology laboratories at the Karolinska Institute in Sweden and at Harvard and the Dana-Farber Cancer Institute appear to have solved this problem by harnessing biological molecular machine systems to make large quantities of very high quality single-stranded DNA oligonucleotides. A hat tip to KurzweilAI.net for reprinting the news from the Karolinska Institute “New method of mass-producing high-quality DNA molecules“:
The new method is versatile and able to solve problems that currently restrict the production of DNA fragments.
“We’ve used enzymatic production methods to create a system that not only improves the quality of the manufactured oligonucleotides but that also makes it possible to scale up production using bacteria in order to produce large amounts of DNA copies cheaply,” says co-developer Björn Högberg at the Swedish Medical Nanoscience Center, part of the Department of Neuroscience at Karolinska Institutet in Sweden.
The process of bioproduction, whereby bacteria are used to copy DNA sequences, enables the manufacture of large amounts of DNA copies at a low cost. Unlike current methods of synthesising oligonucleotides, where the number of errors increases with the length of the sequence, this new method according to the developers also works well for long oligonucleotides of several hundred nitrogenous bases.
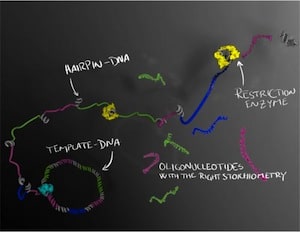
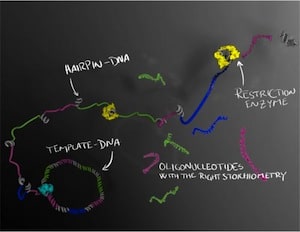
The DNA molecules are first formed as a long string of single-stranded DNA in which the sequence of interest is repeated several times. The long strand forms tiny regions called hairpins, where the strand folds back on itself. These hairpins can then be cut up by enzymes, which serve as a molecular-biological pair of scissors that cuts the DNA at selected sites. Several different oligonucleotides can thus be produced at the same time in a perfectly balanced combination, which is important if they are to be crystallised or used therapeutically.
The research “Enzymatic production of ‘monoclonal stoichiometric’ single-stranded DNA oligonucleotides” was published in Nature Methods [abstract]. A full text PDF can be downloaded from the publications page of Dr. Björn Högberg’s web site. In their “monoclonal stoichiometric’ (MOSIC) method synthetic DNA is cloned into bacteria and individual clones are verified by DNA sequencing to be perfect, and then used as templates for amplification (a much more accurate process than the original solid-state synthesis). The efficiency of the method and the purity of the oligonucleotides produced was demonstrated by producing several types of DNA nanostructures, including the crystallization of a tensegrity triangular structure folded from 7 individual oligonucleotides. They were also able to make 378-nucleotide long oligomers at a cost 15 to 30 times less than chemically synthesize oligonucleotides. The authors note:
The MOSIC method therefore opens a path to construct DNA nanostructures using ODNs much longer than the standard oligonucleotides (<40–60 nt long) normally used for these types of applications. MOSIC could be used to produce designs that are a hybrid between the recently reported single-stranded tile and scaffolded-origami methods.
Perhaps this advance will lead to the construction of more complex and capable DNA nanomachines. Perhaps DNA nanotechnologists will be able to build mechanisms to crudely position molecular building blocks?
—James Lewis, PhD