“The various levels of electrical signal from the sequence of a DNA strand pulled through a nanopore reader (top) corresponds to specific DNA nucleotides, thymine, adenine, cytosine and guanine (bottom).”
We recently noted the contribution of nanotechnology-based DNA sequencing methods to research and to the emerging field of personalized medicine. Another major step along this path was taken more recently by combining a mutated protein pore with a DNA polymerase molecular motor. A hat tip to ScienceDaily for reprinting this University of Washington news release written by Vince Stricherz “Tiny reader makes fast, cheap DNA sequencing feasible“:
Researchers have devised a nanoscale sensor to electronically read the sequence of a single DNA molecule, a technique that is fast and inexpensive and could make DNA sequencing widely available.
The technique could lead to affordable personalized medicine, potentially revealing predispositions for afflictions such as cancer, diabetes or addiction.
“There is a clear path to a workable, easily produced sequencing platform,” said Jens Gundlach, a University of Washington physics professor who leads the research team. “We augmented a protein nanopore we developed for this purpose with a molecular motor that moves a DNA strand through the pore a nucleotide at a time.”
The researchers previously reported creating the nanopore by genetically engineering a protein pore from a mycobacterium. The nanopore, from Mycobacterium smegmatis porin A, has an opening 1 billionth of a meter in size, just large enough for a single DNA strand to pass through.
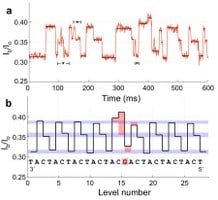
To make it work as a reader, the nanopore was placed in a membrane surrounded by potassium-chloride solution, with a small voltage applied to create an ion current flowing through the nanopore. The electrical signature changes depending on the type of nucleotide traveling through the nanopore. Each type of DNA nucleotide – cytosine, guanine, adenine and thymine – produces a distinctive signature.
The researchers attached a molecular motor, taken from an enzyme associated with replication of a virus, to pull the DNA strand through the nanopore reader. The motor was first used in a similar effort by researchers at the University of California, Santa Cruz, but they used a different pore that could not distinguish the different nucleotide types.
Gundlach is the corresponding author of a paper published online March 25 by Nature Biotechnology [abstract] that reports a successful demonstration of the new technique using six different strands of DNA. The results corresponded to the already known DNA sequence of the strands, which had readable regions 42 to 53 nucleotides long.
“The motor pulls the strand through the pore at a manageable speed of tens of milliseconds per nucleotide, which is slow enough to be able to read the current signal,” Gundlach said.
Gundlach said the nanopore technique also can be used to identify how DNA is modified in a given individual. Such modifications, referred to as epigenetic DNA modifications, take place as chemical reactions within cells and are underlying causes of various conditions.
“Epigenetic modifications are rather important for things like cancer,” he said. Being able to provide DNA sequencing that can identify epigenetic changes “is one of the charms of the nanopore sequencing method.”
The ability to identify epigenetic modifications (mostly methylations of specific nucleotides) is indeed a plus, although we can hope that with further development the technology will be able to read DNA sequences far longer than 42 to 53 nucleotides. Because repeating sequences are prevalent in the human genome, the ability to do long reads is very important.
—James Lewis, PhD

