Summary
In this session, Brain Kennedy from Buck Institute presented his assessment of the state of aging research in academia and the non incentivized academic research that could dramatically advance progress. After that, Lynne Cox introduced two progress opportunities she identified – new drug discovery approaches focusing on polypharmacology, and in silico systems modelling of aging. Following that was a last presentation from Joris Deelen, in which he presented ideas on how to get already discovered aging biomarkers into the clinic, as well as a novel approach to utilize genetics of long lived people to move the field forwards.
Presenters

Brian Kennedy, Buck Institute
Dr. Kennedy earned his PhD from the Massachusetts Institute of Technology and is well known for his work during his graduate studies with Leonard Guarente, PhD, which led to the discovery that sirtuins (SIR2) modulate aging. He performed postdoctoral studies at the MGH Cancer Center associated with Harvard Medical…

Joris Deelen, Max Planck Institute for Ageing
Joris Deelen is leading the research group on Genetics and Biomarkers of Human Aeging at the Max Planck Institute for Aeging Research. The group studies the genetic mechanisms underlying healthy ageing in humans by investigating the effect of genetic variants that are unique to long-lived families on the…

Lynne Cox, Oxford University
Lynne Cox is the George Moody Fellow and Tutor in Biochemistry at Oriel at the University of Oxford. Her lab studies the molecular basis of human ageing, with the aim of reducing the morbidity and frailty associated with old age. Her lab studies progeroid Werner’s syndrome (WS), where cell senescence and early onset of many…
Presentation: Brain Kennedy
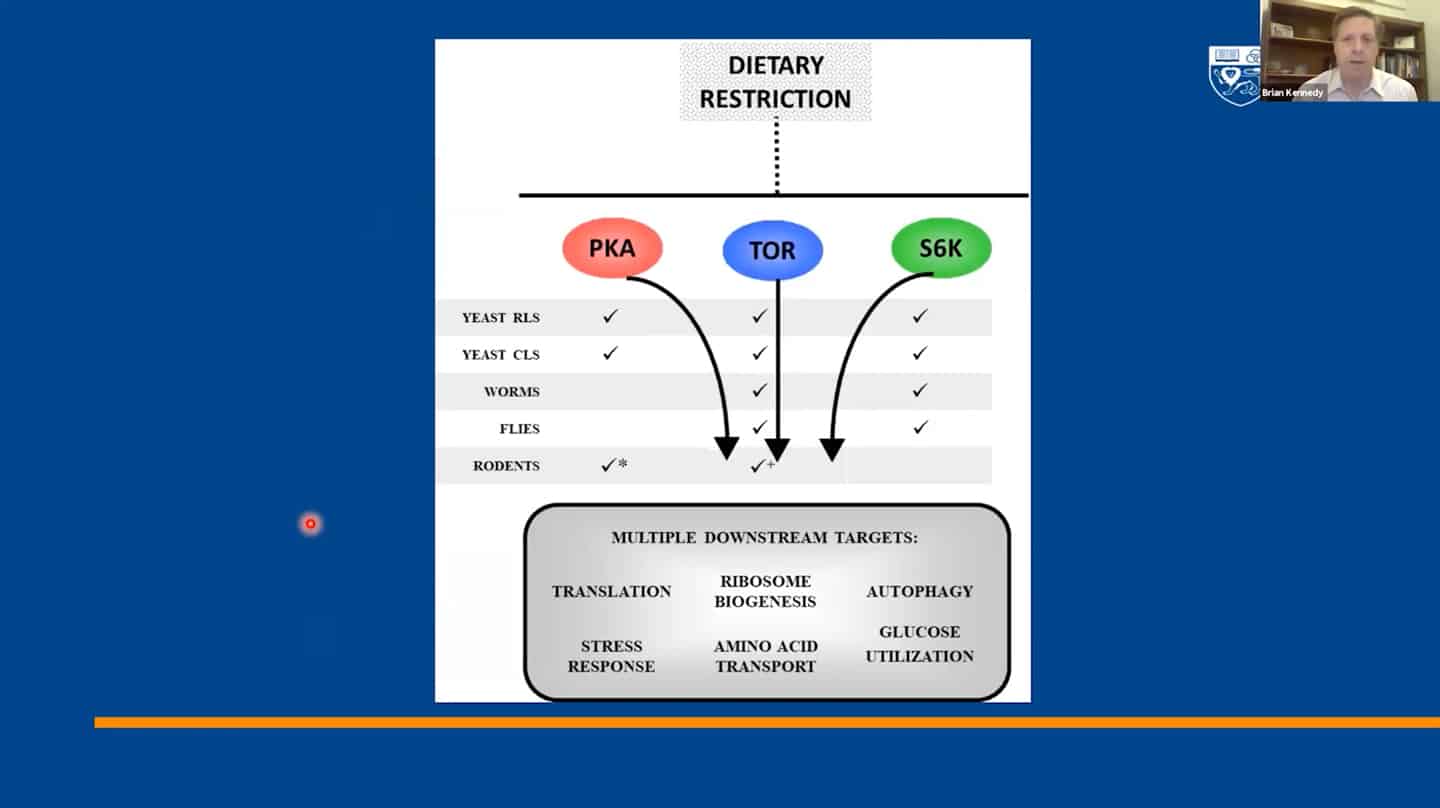
- We identified 300+ genes via basic research on invertebrates which served as a basis for most of the current research, but now basic research and model organisms are kind of overlooked in terms of funding and focus. It’s great that private money is entering the longevity space, but that seems to generate this unfortunate idea that basic research is something that’s no longer needed and not a priority. And yet we don’t even know how the 300+ genes regulating aging in yeast interact with each other, which ones regulate which pathways, how they coordinate aging, etc. Single-cell organisms like yeast may be the only way to answer this question.
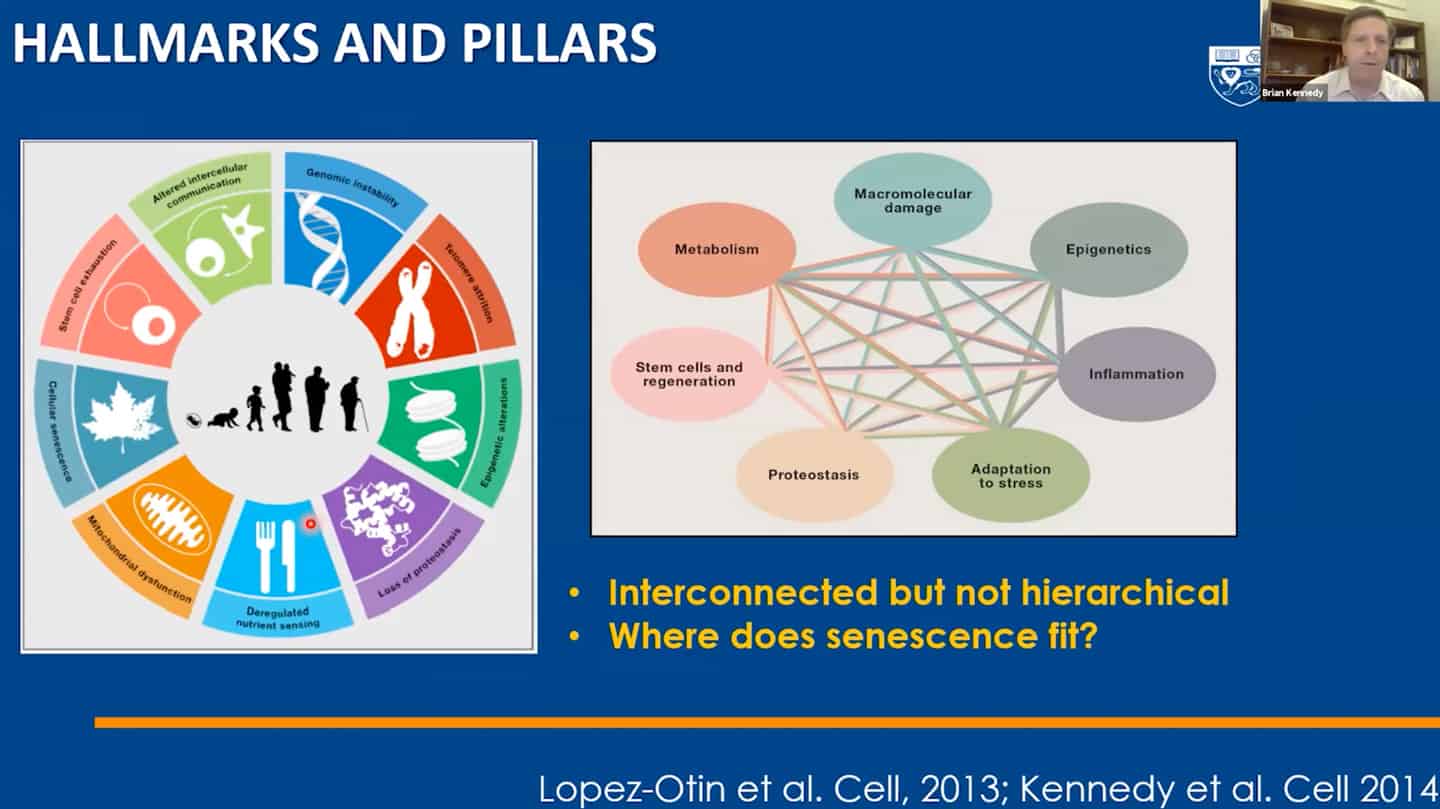
- We managed to categorize aging in pillars of aging / hallmarks of aging, but we still don’t understand the interaction between hallmarks of aging – that’s another example of open questions that are not being addressed.
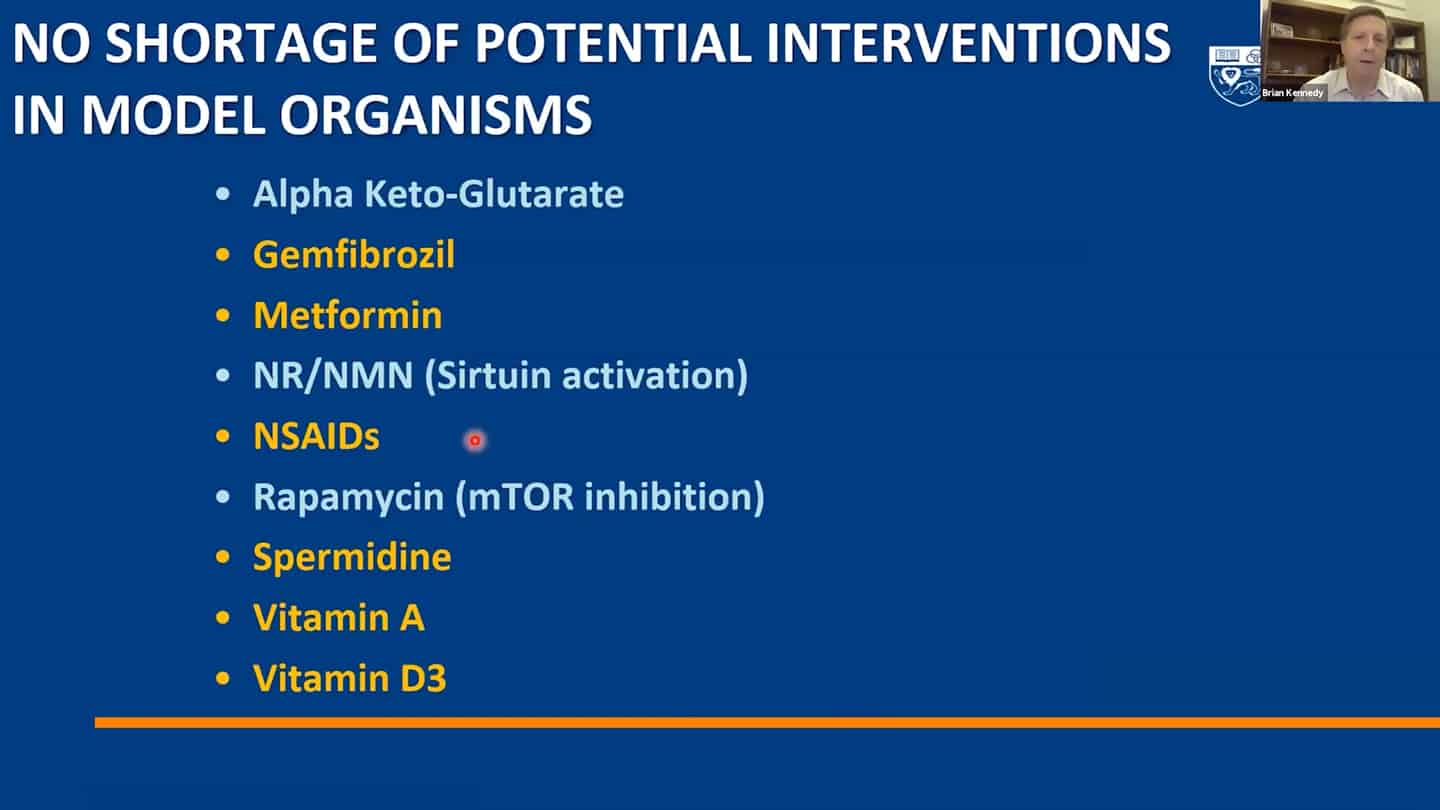
- A lot of small molecules are now funded by companies, but a lot of them are still not and we are continuously discovering more.
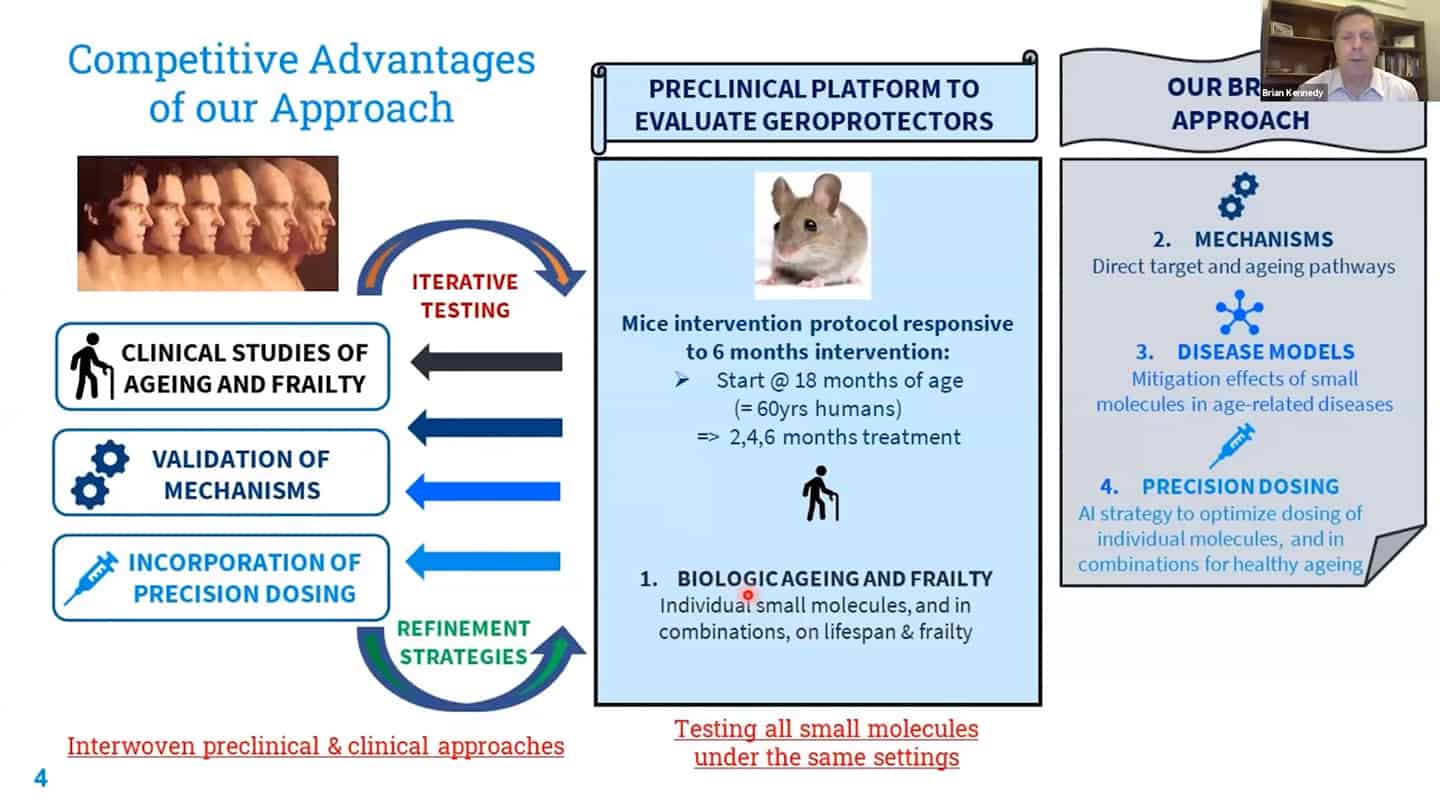
- Roadmap to get to humans.
- Three different strategies for companies on how to get to humans.
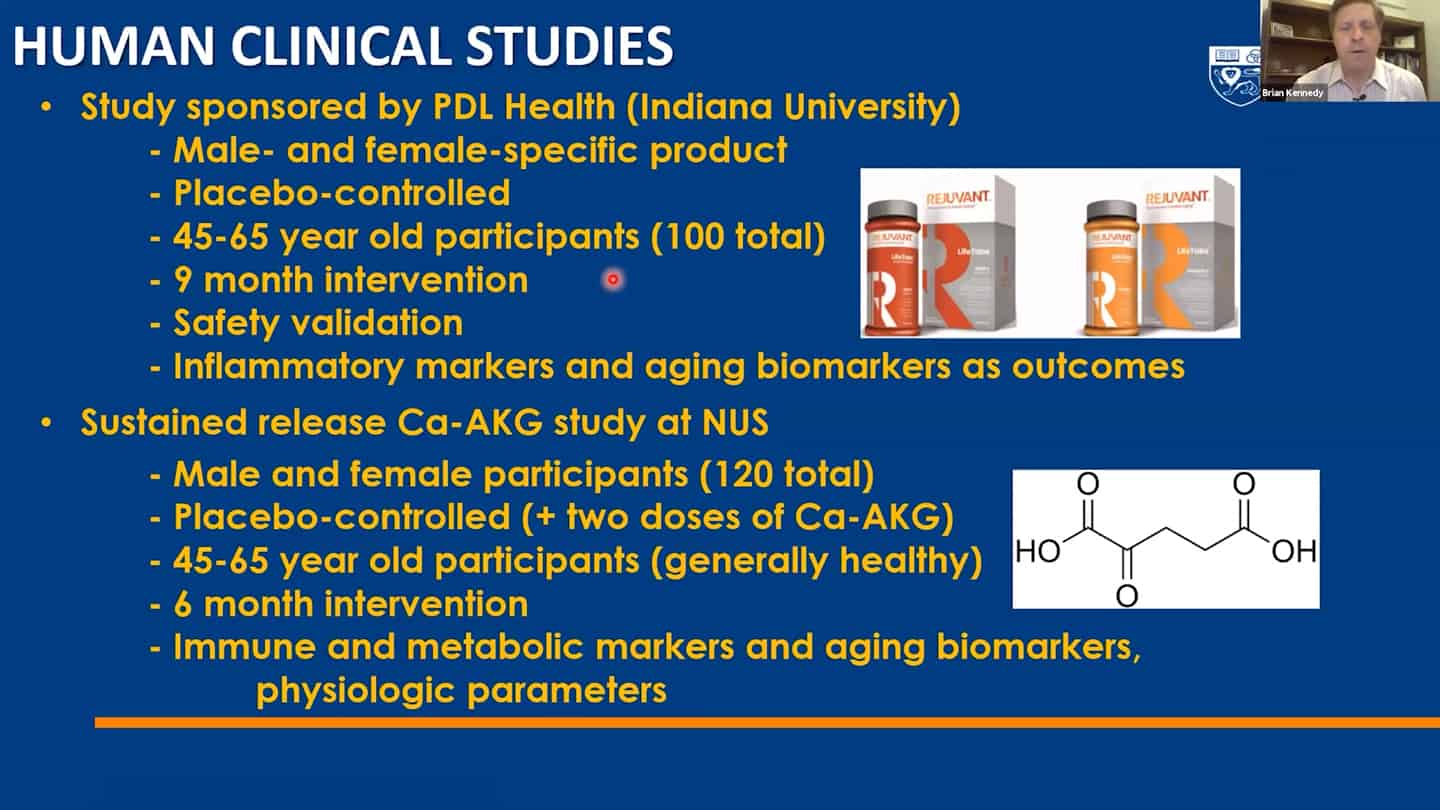
- We are working on recalibration of mouse studies, so that they are aligned with these shorter-time biomarker studies in humans, so we can work back and forth between humans and mice using validated biomarkers.
- Example of a study we’re running. Trying to test up to 10 of these molecules so we can contrast and compare how different interventions affect aging.
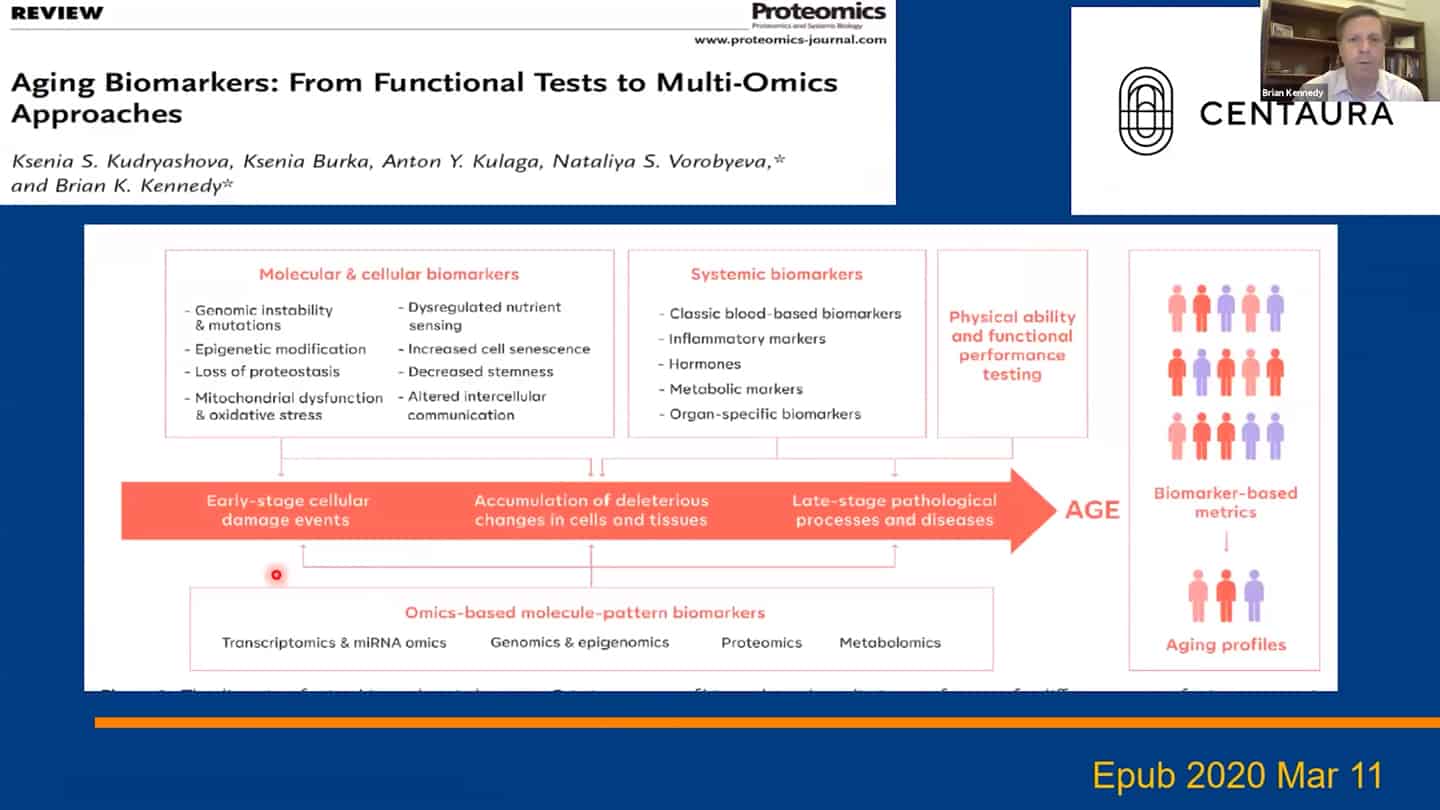
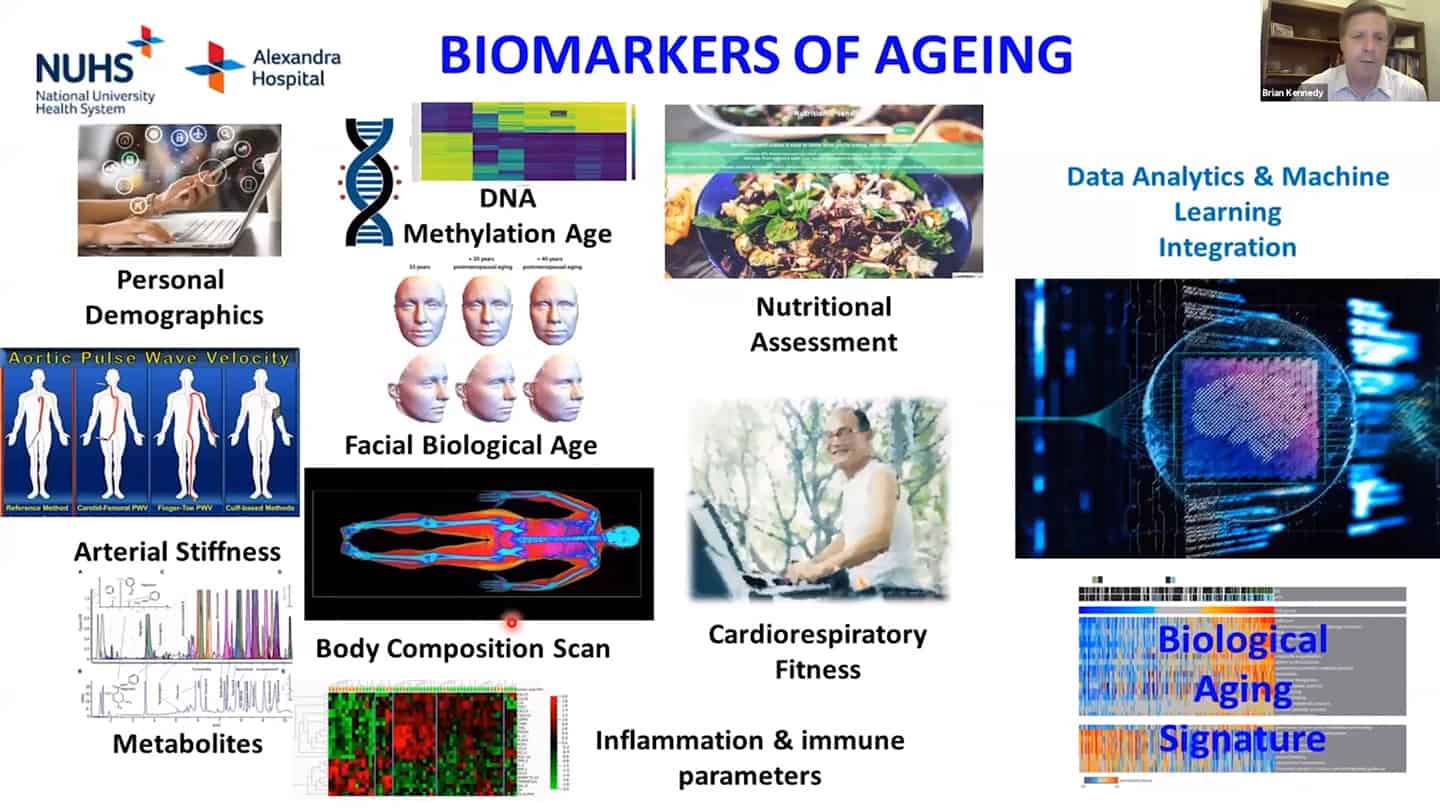
- We are using a number of different biomarkers for these studies, but there is the problem that we don’t know how aging biomarkers interact with each other.
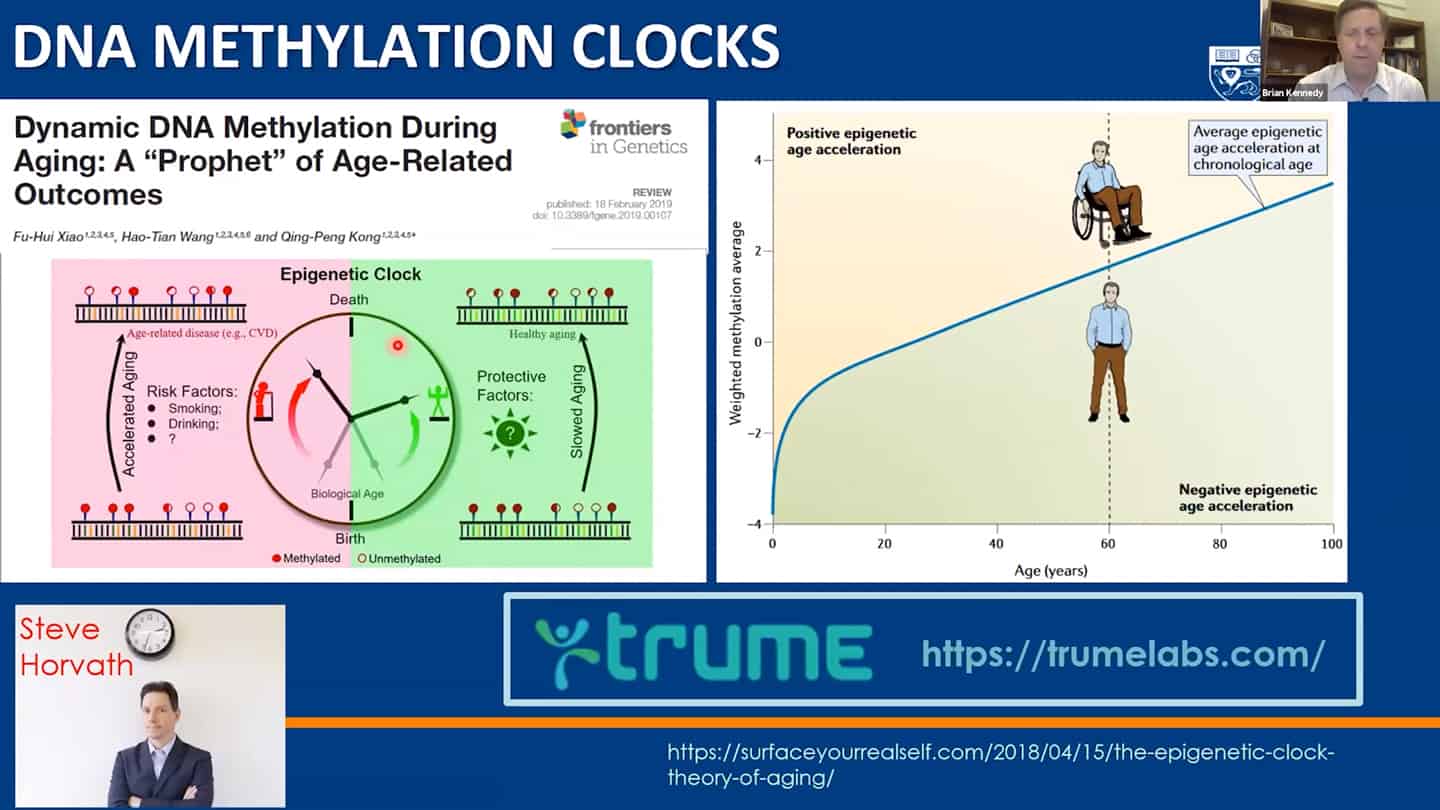
- Methylation clocks are commonly used, but even there are different methylation clocks, with different parameters, etc.
- We need to combine these interventions together, as well as measurements of the hallmarks together and validate what works in humans.
Q&A
What is the lowest hanging fruit in your opinion?
- What do we mean when we say network regulating aging? How do we directly find out when we modulate a certain pathway, how do we primarily assign the role in aging? If it is just maintaining the network, what does it mean from a biological perspective? We see a lot of claims that this intervention affects inflammation, stem cells, etc., but what does it mean in terms of primary effects of this intervention and how does that get translated into the preservation of the network?
- Yeast research is great for finding limitations of waves of interventions: Especially for figuring out the limitations and waves of limitations that will need to be addressed after we are able to address the current limitations we are aware of with our current understanding (that is being translated now). This was revealed in earlier research in yeast – when we knocked out all the genes we thought affected lifespan positively and it worked, we ran into another set of different limitations in that yeast with prolonged lifespan. And we wouldn’t know about it if we didn’t conquer the first wave limitations first. It’s likely that there will be such a second wave of limitations and barriers in humans as well.
Key discoveries were made in academia that the private sector couldn’t fund:
- There are many more known unknowns and unknown unknowns. So we cannot forget that and should try to get more people working on basic science in aging, and let serendipity run its course instead of investing $100M specifically into Alzheimer’s research. More and more people are focusing on translational research, which is good on one side but there’s a risk on the other that we lose long-term discoveries that could have even more value.
Focus on commercialization might actually have an upside if done well:
- Hopefully fitting analogy is what happened with the computer science field – scientific discoveries led to the internet, which led to the dotcom boom, which led to more people going into academia and doing basic computer science research. There was an international effort to set up standardized tooling and an interoperability scaffold for the entire field, allowing all types of organizations – both public and private – to build on top of it – that should be the goal for the longevity field as well.
Presentation: Lynne Cox
- Agree with Brian that we need to tie things together in the hallmarks of aging space, we think we know quite a bit about it and we need more, but there are some other things that we simply don’t know enough at all.
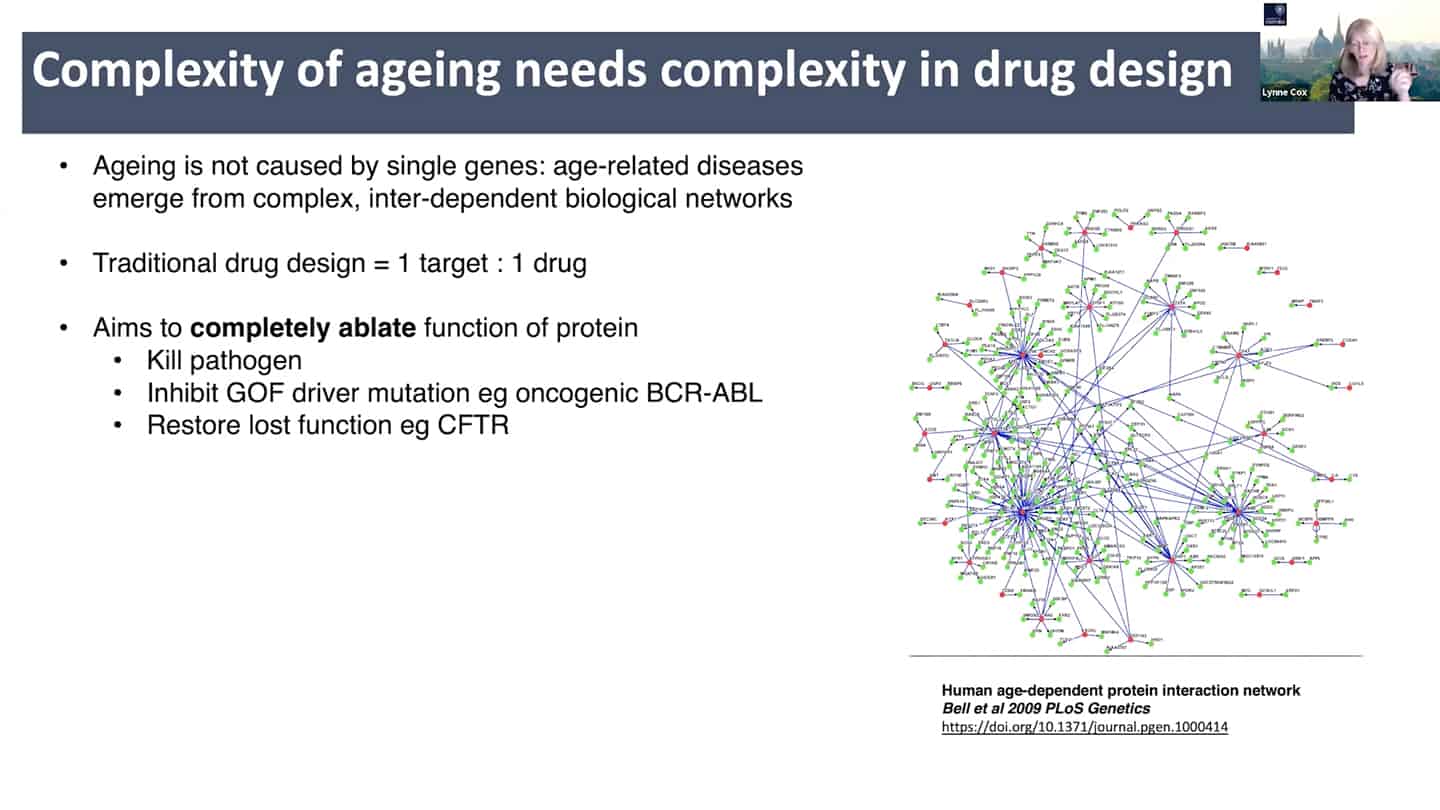
- New drug discovery approaches are needed. Aging is a complex inter-dependent and emergent network. Traditional approach is 1 target = 1 drug, where you try to annihilate the target completely to totally ablate it’s function. This does not work well with aging because of the interconnectedness and redundancy in aging pathways. There is probably broad agreement on this, but it is insufficiently stressed, perhaps because ultimately getting different (perhaps multi-target) interventions past FDA/EMA has historically been unusual outside of oncology and diseases like tuberculosis.
- See papers: Limits of the classic 1 knockout at a time methodology, Genetics of extreme human longevity to guide drug discovery for healthy ageing
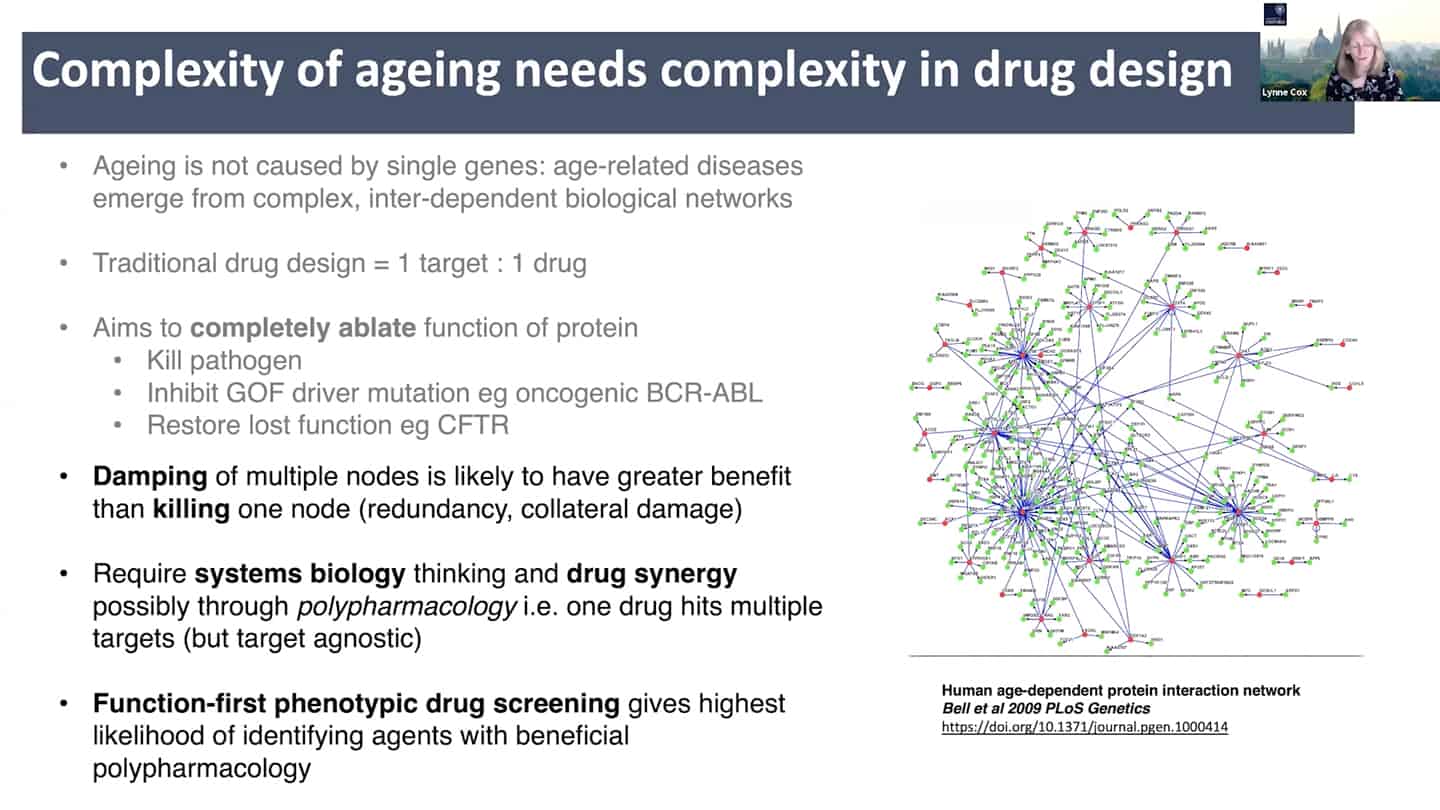
- Current pharma philosophy of inhibiting target completely is like a hammer to fix a clog on a wristwatch, we need to find better solutions like modulating and damping down multiple nodes, rather than killing them completely, akin to fine tuning and tweaking wristwatch with tweezers instead of a hammer.
- This requires systems biology and drug synergy approaches – polypharmacology. Instead of 1 target = 1 drug, develop X targets = 1 drug approach, where one drug hits multiple targets and is target agnostic. And that approach requires function first phenotypic drug screening.
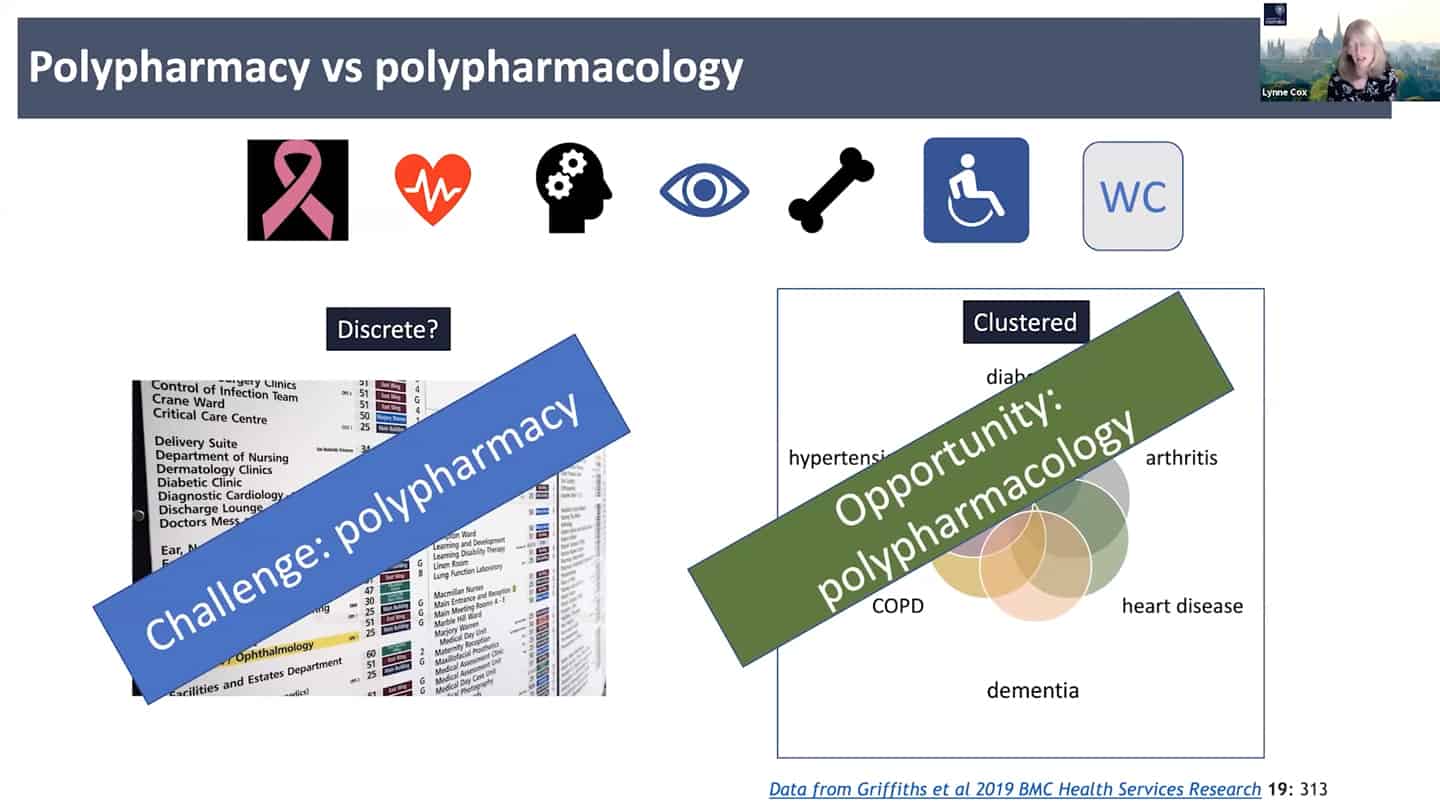
- Right now the standard approach is polypharmacy, for every problem a different expertise and clinic, instead of looking at the common denominators. We need to look at the shared clustered reasons, like aging, where we can hit the core of it all.
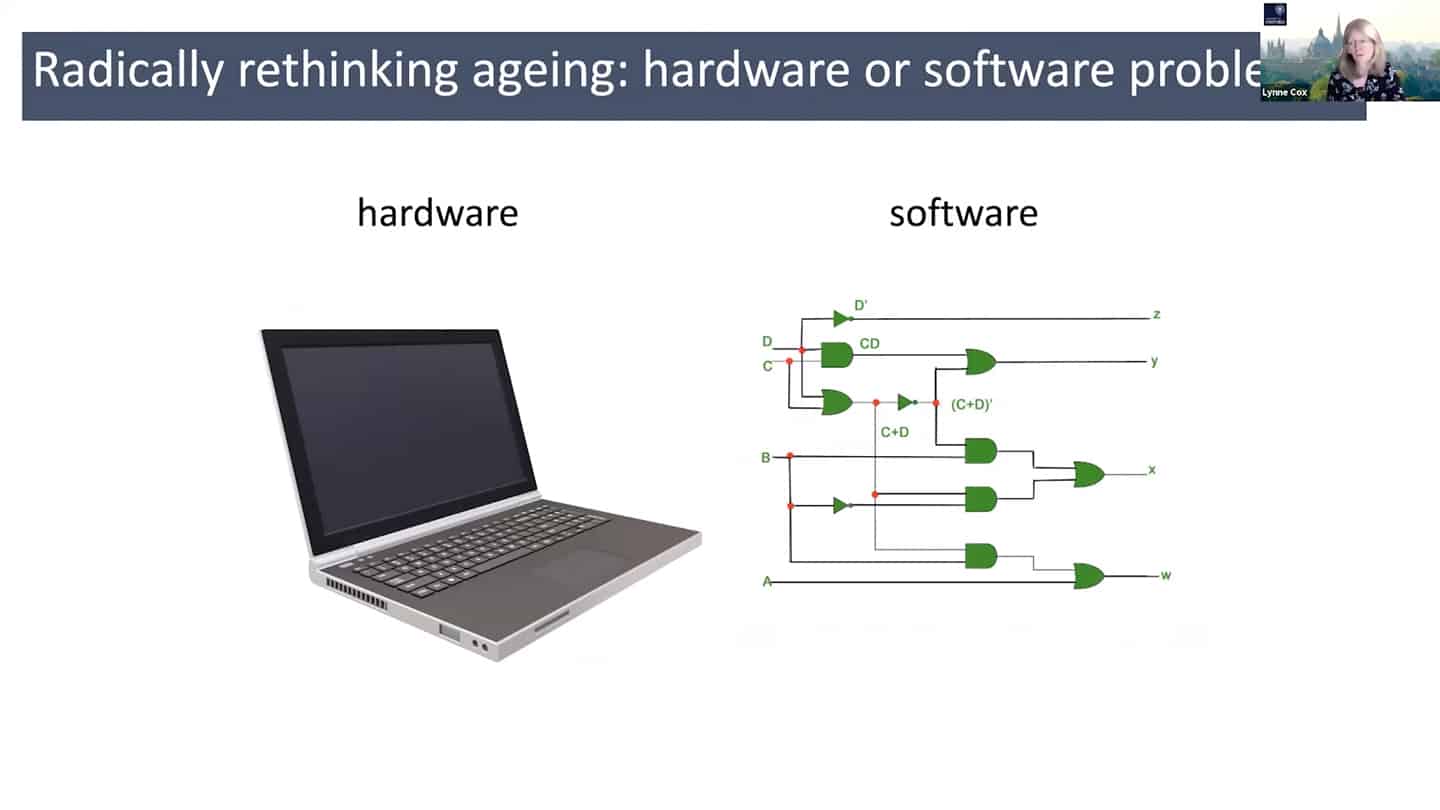
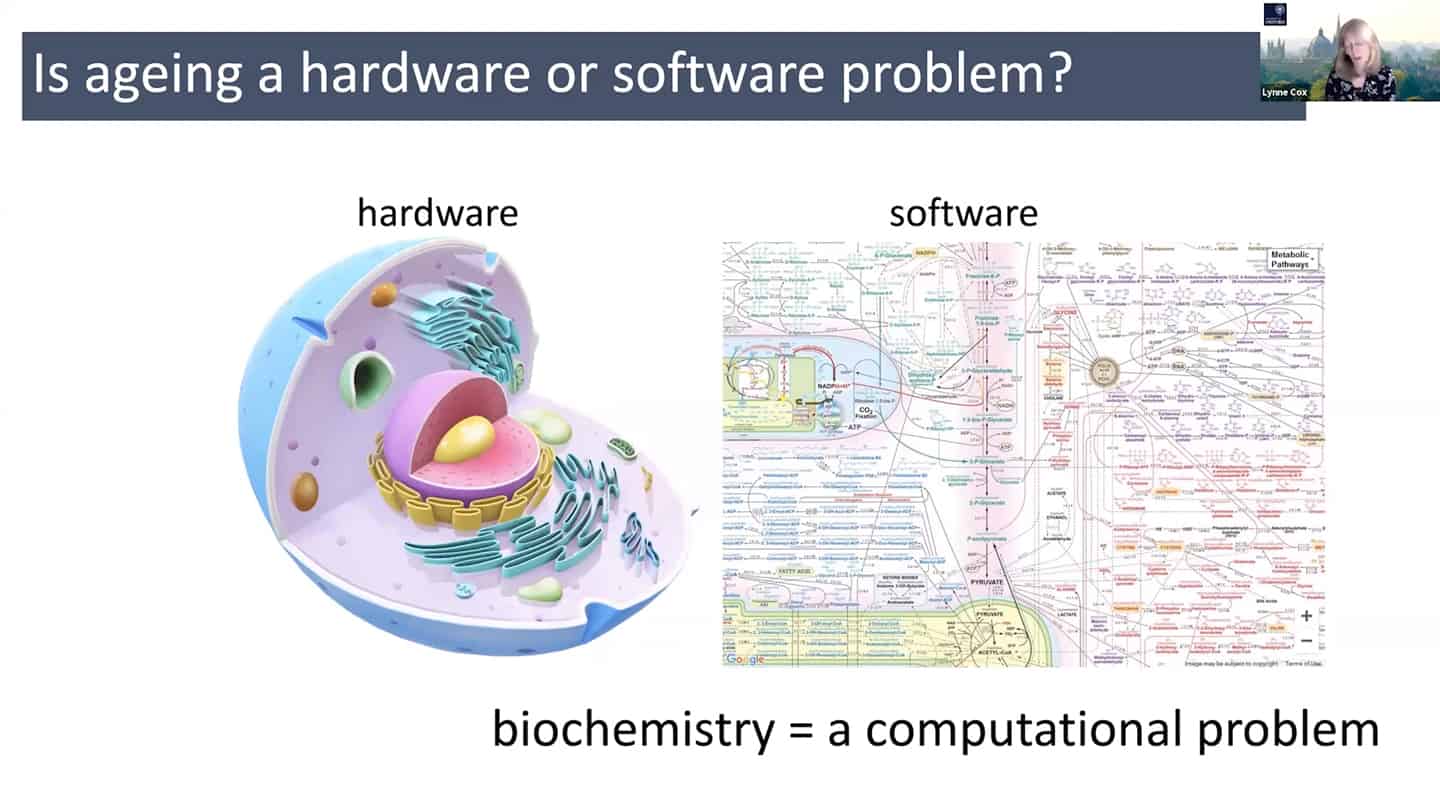
- In order to do all that, we need a different understanding of aging – maybe radically rethinking aging as a hardware and software problem could be an useful framework.
- In quite a reductionist sense, hardware is essentially a cell, organelles or macromolecules, while the software is the way that those things talk to each other, how the information flows through the system through biochemical pathways. And so we can think about biochemistry as a computational problem, and maybe as we accumulate enough data, we could plug it into a model and start to mimic aging in silico.
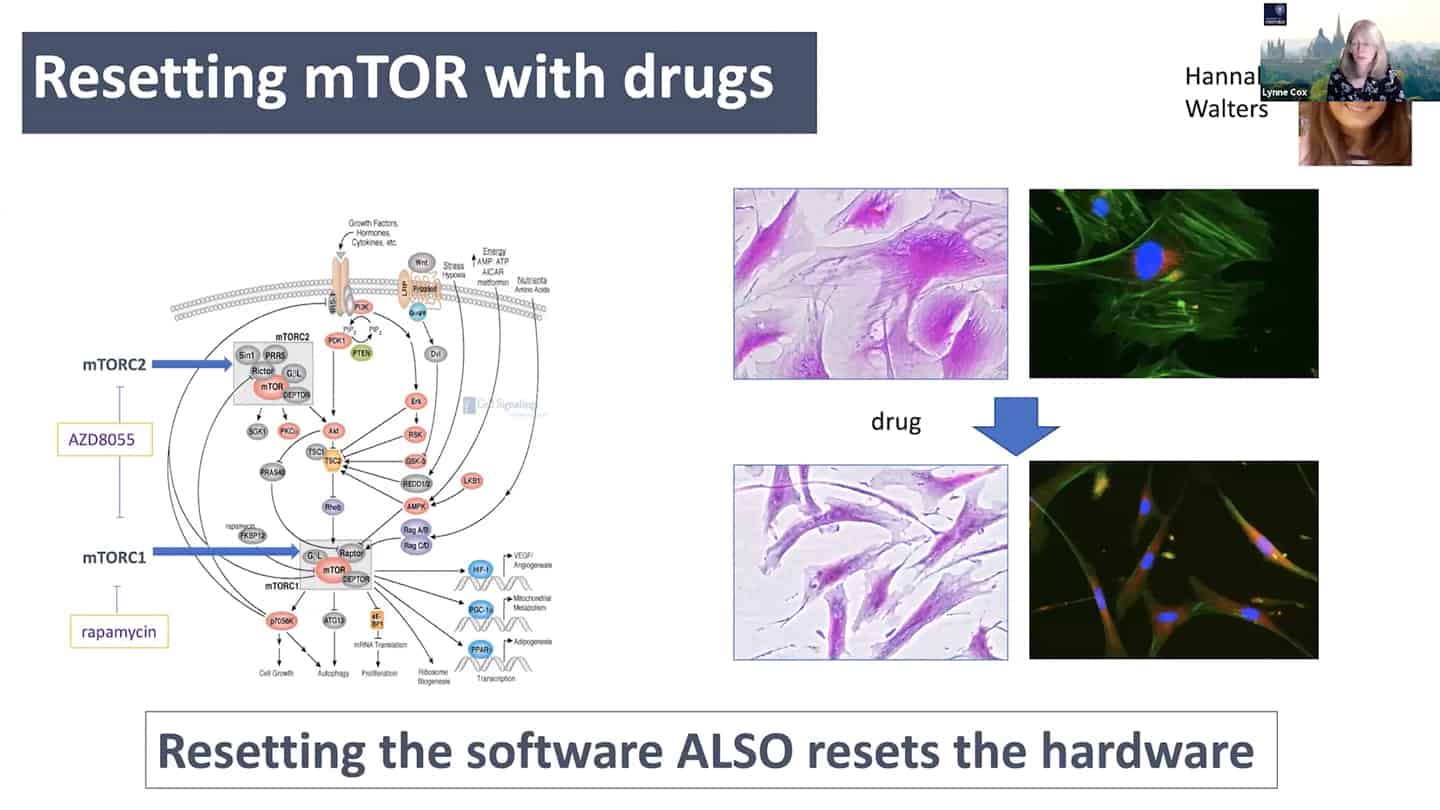
- mTOR is a good example – we know that young cells have correct mTOR programming that switches on and off, but in old senescent cells it is switched on all the time. So we can start to think about it in a computational way as a logic gate. In the young cells, it is a correctly working AND gate, and in the old cells, it is incorrectly working OR gate. So all we have to do is to go there and debug the software.
- As a proof of concept, we tried to reset the mTOR with a pan-mTOR inhibitor to see whether we can reset the software, and interestingly resetting the software did reset the hardware as well.
- We know that we don’t know how these genes interact with each other, how the cell components in these systems interact with tissues, organs, and systems, with microbiome, and even more how environmental factors affect all of that, both generationally and intergenerationally. And so what we need are biomarkers, we need to know what’s going on, we need full biochemical workup, systems modelling, and we also need big data. Not just biology -omics (geromics for gerontology), but also other factors like socioeconomic data (e.g. Open Life Data Framework).
Q&A
To your knowledge what are best in silico models applicable to what you just said? (e.g. compbiomed.eu, Karr lab, Ideker lab,…)
- There are a few online models trying to build a virtual cell, but essentially I don’t think they’re based on enough biochemical data to make any sense. One is based in Germany, but it doesn’t model the complexities we know exist (where we can take one variable and see all the downstream effects, right now you can take one variable and you can read maybe one or two factors), which is necessary for progress. Another group is in Newcastle that is also doing this, but the complexity is just not there yet, probably due to the quality of material and data used for the modelling.
- References:
Are there areas where systems pharmacology is limited by tools, either for characterization of experimental intervention? That’s one way how science progresses – basic scientists say this is a known unknown, and something we would like to be able to measure or experimentally intervene, we can’t do it now, and then ask people in physical sciences or engineering to develop a tool with a set of performance characteristics.
- A tool for generation of new chemical spaces: Right now we are using natural product or general chemical libraries, but there is an opportunity for combinatorial libraries made of drug-like fragments. And we need tools in order to generate those. Essentially a tool to create new chemical space agnostic to the way you develop it, but the readout is simply whether you have a phenotypic change based on the activity of products that the particular component has managed to generate.
- Better target agnostic readouts on granular level: Because the readouts are so complex and we want to be target agnostic, the target should be for example a senescent cell and the readout should be whether the senescent cell is not senescent anymore after.
Talk – Joris Deelen
Biomarkers and how to move them to the clinic
- Benchmark and compare aging biomarkers with existing ones: We should try to use all the already identified aging biomarkers and test them along with biomarkers that are already used (cholesterol, triglycerides,..) and bring them to the clinic this way, and see if it can replace the things that are already used in the clinic.
- Harmonize clinical trials and blood biomarkers used in mice and humans: Not many biomarkers that are coming from the model organism are actually used in the clinic. What might help is testing more from the blood of mice and then harmonizing it with human blood (we usually do blood tests with humans, but with mice we take all kinds of tissues, which are not possible to collect from humans for wide use). And then testing an intervention that works in mice on humans and see exactly how it differs. Harmonize mice clinical trials to look more like human clinical trials, to mimic the human situation, which could hopefully make them a bit more translatable.
Utilize genetics of long lived people to move the field forward
- Go from long lived humans to animals instead of from animals to humans: A lot of research comes from animal models, which works well, however we sometimes do not see the same effect in humans. And in long lived humans, it’s really hard to find common genetic mechanisms between all these people that would explain why they managed to get so old. There might be another way to use the data from long live humans though. And that is developing interventions to those possible identified targets that we’re not sure about, and then test them in mice. So instead going from animals to humans, go from long lived humans to animals.
- Dampen rather than completely block: For example rapamycin completely blocks mTOR1, but we don’t see mTOR1 being completely blocked in long lived humans. There’s more of a dampening rather than shutting off completely, so we should look for interventions that are mimicking that.
- Create a study and a database with all different biomarkers and compare them: One actionable thing we could do would be to collect all different types of biomarkers and test them in a study. They have never been tested in the same study, and in the right study. Having such a database to make a decision about specific biomarkers and omics would be helpful.
- Develop markers and tooling enabling cheaper testing on a massive scale: Some of these markers are very costly though, so another action point is developing markers that are much cheaper. So we can get much more data and test the markers on larger scales. So doctors and researchers can use them and agree on them, because they are proven to be valid. If there isn’t any database like that yet, we should come up with one.
Q&A
We’ve been looking at biomarkers in a very limiting way:
- There is a new biomarker used to diagnose Parkinson’s based on lipids in sebum. Incredibly cheap and easy way of doing it, based on one nurse’s ability to smell Parkinson’s patients. Similar to that, we as a field could be a bit more adventurous about the types of things that we consider as a biomarker.
Collecting samples from humans is impractical unless it’s blood, what are some other sample modalities?
- Maybe non or minimally invasive things like skin biopsy, urine, saliva. Problem is that it’s hard to get high throughput data for the less used modalities. That might be an opening for new tools that are able to do that. To bring something into the clinic, it needs to be high-throughput and easily measurable, and also standardized so we can compare between different studies. So we can do 1000s of samples for large studies and then clinical use. That is a challenge that we should be working on, so we can get bigger databases. There’s a focus on discovering biomarkers, but not much focus on making those measurements affordable and available in high-throughput fashion.
What should government funded research be doing with future centenarians to track them now? What does Nir Barzilai wish he would have done before starting the original study on centenarians with current tools?
- Nir Barzilai, Thomas Perls, AFAR, Regeneron are together starting a project with the aim to recruit the next 10000 supercentenarians, and get all the data (whole genome sequencing, electronic medical record,…) from them. They are also recruiting their offspring, because when looking at supercentenarians, you are never sure whether you are looking at what enabled them to live that long, or what will kill them in a year, because they’re at the end of their life. But offsprings should be a great treasure trove of the right data (for example IGF and HDL levels, which proteins they have even in youth and are inherited, or which special proteins they have, etc.).
Do methylome clocks on the offspring of supercentenarians look better than the age matched controls?
- They are just studying that, and it seems that they actually fall between the supercentenarians and controls. But supercentenarians are older, so the question is which methylation patterns are inherited – it is possible that the methylation patterns are inherited, so there’s a lot of work to do on the biology of methylation and not only the clocks.
Do supercentenarians have a shared mechanism or are there multiple mechanisms how to get there?
- We don’t know yet, but we might be able to see soon with the increasing amount of data that we run through models. The more data we have the more needles in the haystack. However even now, if we put the genetic differences among supercentenarians into pathways, they are really telling – insulin IGF pathway, mTOR pathway, AMPK pathway, sirtuin pathway. So it’s probably all there, we just need to know what to ask. Pathway analysis is a key to this, we cannot look at single proteins as biomarkers, because there are not statistically significant differences most of the time. But when we look at it on the systemic level, and take 50 proteins that are involved in the exact same biochemical pathway and they all shift in the same direction, then the pathway becomes really significant as a whole.