Presenters

William Shih, Harvard University
William is overseeing an effort to apply Synthetic Biology approaches to the development of self-assembling DNA nanostructures and devices for use in biomedical applications. To achieve structures of even greater complexity, his laboratory is pioneering methods for hierarchical assembly of these particles into three-dimensional networks with site-specific control over chemical functionalization and mechanical actuation…

Lulu Qian, California Institute of Technology
Designing and constructing artificial molecular systems that exhibit programmable behaviors such as recognizing molecular events from their environment, processing information, making decisions, taking actions, learning, and evolving….
Presentation: Single-molecule Detection and Identification via DNA Nanotechnology
- Criss-cross polymerization technology – an up and coming method to supplement things like ELISA and Mass spectrometry.
- DNA origami – a robust process for creating DNA structures using scaffold strands and free floating oligonucleotides. However, it can only be used for structures <100 nm in size. To work with larger structures, criss-cross polymerization can be used.
- Why would we want to create a large DNA origami structure? One reason would be to amplify an analyte signal to billions of times its size. Detecting DNA that exists in zettamolar quantities in serum – maybe the genome of a pathogenic virus at the very start of its infection. To detect these, a two step process is used. The first is to detect the target sequence and convert it into an artificial record, called a nano-bean. The second step is to convert each nano-bean into a micro scale beanstalk, a large scaffold entity which can be detected easily. The key is to have a beanstalk formation process which is only ever initiated by the target nano-bean.
- Crisscross-cooperative growth uses DNA fragments built to only polymerize with each other on 3-4 base pairs. As the scaffold grows, the DNA is locked in place by making contact with successively further DNA fragments, creating a weave of DNA where base pairs further from the center are matched with each other. The scaffold can only grow one specific fragment at a time due to binding efficiency within the scaffold.
- Free binding of these DNA fragments requires high activation energy, preventing spontaneous scaffold formation. By engineering a seed, we can initiate a binding cascade that generates a long scaffold of woven DNA.
- The scaffold then grows to a size that becomes visibly large.
- These ribbons can be programmed to twist together and form tubes.
- Converting this principle into a mechanism to identify specific sequences is tricky. One method may be to use six initiator DNA probes to fold the target sequence into a DNA origami, becoming the structure of the seed described earlier. The probes then form the base for ribbon formation, and can generate a binding cascade.
- This mechanism has applications for all-or-nothing growth of microstructures, fast algorithmic assembly, and rapid zero-background amplification for ultradetection
- The next mechanism is a caliper that can measure extremely small objects
- In this example, a peptide of unknown length has DNA attached at various points – one long strand at one end, and shorter strands attached to lysines at varying lengths. The caliper consists of two microbeads held in place by a dual beam optical trap, tethered to a long DNA strand of which a section consists of only a single strand of DNA. The single-strand portion grabs both the long DNA probe at the end of the unknown peptide as well as one of the small probes attached to a lysine. When this happens, the beads close together slightly and the difference in length can be measured.
- Not shown on the slide – imagine the single strand portion is purple at the bottom and red at the top. The small fragment will always bind to the top of the single stranded portion, and the purple fragment will always bind to the bottom. The caliper therefore generates an inverse measurement where small peptide lengths induce a large compaction while large peptide lengths induce a small compaction.
- By measuring repeatedly, a histogram can be created and the mean value generated should correlate to the peptide length. Calibration curves generated using this process have perfect correlation except extremely small analytes which likely have stearic problems, and extremely long analytes that are larger than the length of the loop.
- This method can resolve distances of individual amino acids on a peptide chain, giving a sequence fingerprint of a protein.
- The next step is to get sub-angstrom measurements from a single measurement and increase measurement flowthrough.
- In the future, we may be able to measure distances on a folded protein with this caliper, and by measuring millions of distances in various directions produce an accurate 3d model of what the protein structure.
- Potential applications are low-cost, enzyme-free, ultrasensitive digital diagnostics and single-macromolecule structure identification. Near term challenges are to increase the speed and resolution of measurements
- For the midterm future, we may be able to diagnose disease using molecular robots by integrating a large number of inputs
Q&A
At these levels of detection, are they not too sensitive for medicine? You can find a few molecules of all sorts of things.
- There are a lot of analytes for which this may be overkill, but this can still be useful via serial dilution. However, for me this is exciting because there are a lot of analytes which are rare, like viral infections, and we want to know at the earliest point whether we are infected.
Do you think this would be a good candidate for ARPA-H program?
- Absolutely, I think these kind of technologies let us open up a treasure chest of information about our health. Frozen serum and blood samples are just waiting for new technologies to be applied to stored samples.
Do you have an idea for a new model for experimentation, such as the ARPA model?
- I do like the idea of investing in the person, not the project. Having a balanced portfolio of larger opportunities is good but do not diminish the traditional lab investment. Innovation comes from the person level.
How can the calipers cope with molecules that can undergo lots of conformational change?
- We might use calipers where we bind the molecule in such a way that it becomes the loop.
Presentation: Molecular Pattern Recognition in DNA-based Artificial Neural Networks
- Molecular pattern recognition in DNA-based artificial neural networks
- Being smart means recognizing a situation correctly and making a good decision
This nanobot can be considered smart for recognizing when to open and deliver the payload within.
- Recognizing things can be tricky
- Neural networks are good at recognizing things. Also, the same network can be used to recognize different patterns.
- It has been hypothesized that pattern recognition occurs at the molecular level in cells.
- One of the simplest types of network model is the linear threshold model, where the output is 1 if the weighted sums exceed some threshold. 10 years ago we developed a DNA based 4 neuron version of this model that can recall a pattern based on incomplete information.
- The model was scaled up from 4 bit to 100 bit. The model was changed to a competitive “winner take all” method. One advantage is that negative weights are not necessary, because only the relative values of weights matter. This simplifies the system and requires fewer molecules.
For winner take all, an output is 1 if and only if the corresponding input is the largest input.
- Adding a layer prior to computing the winner take all weights allows for pattern recognition. If weights are binary, the function is equivalent to computing the smallest Hamming distance between input and weights.
- This method allows for analog values as well. The winner take all circuit can recognize patterns even if they are heavily corrupted.
- A simple chemical reaction network can implement this winner take all circuit. The annihilation reaction allows all strands to compete with each other until there is only one winner left. The catalysis reaction allows the remaining signals to be amplified.
In the context of DNA, a displacement mechanism is used to simulate competition. An invading strand binds at a toehold and then competes with the native strand. If it wins, the native strand is released and signals that competition has been resolved.
Invading strand = Input signal
Native strand = Output signal
Double strand = Gate molecule
- The annihilator function allows different DNA signal molecules to destroy each other until only one winner is left
- Catalysis restores the signal after annihilation.
- In order to force the annihilation reaction to occur faster than the catalysis reaction, longer toeholds are engineered on the annihilator strands than the restoration gate.
- Mixing these molecules in a test tube allows all steps to be performed
- Highly complex identification tasks can be carried out. Sloppy handwriting can be used as an example here.
- 100 sixes and sevens were averaged into a weighted analog profile.
- Each bit corresponds to two DNA molecules that perform the weight multiplication function
- In the database there are over 14k handwritten sixes and sevens. The weighted sum space tells us how easy or hard the winner take all should be. Further away from the diagonal line means easier, closer is harder.
- 2% of the patterns are on the wrong side of the line, making detection impossible
- 8% are close to the line, which makes detection difficult
- 90% are far enough away to recognize correctly. 36 were chosen for experimental demonstration.
- Perfect patterns were recognized, but also patterns with up to 30 bits of deviation from the template were still recognized.
- The process was scaled up using an array
- All 9 digits can be accurately identified
- An online computer simulation of the process was developed
- Undergrads and grads with minimal prior background were able to construct identification methods using this tool
- Potential applications include molecular diagnostics and therapeutics, cell type imaging, or water pollution detection
- A proof of concept demonstration showed identification between viral vs. bacterial infections with just 7 inputs. One challenge is the low concentration of RNA in blood samples – developing a signal amplifier is an important step for diagnostics.
- A challenge for therapeutics is to scale up the number of DNA circuits for a DNA robot and improving the robustness of strand displacement reactions.
- B cells and T cells can be labeled with DNA barcodes based on cell surface markers. This leads to localized reactions rather than reactions between diffused products. The challenge here is to understand the nature of localized reactions between DNA tagged to surface proteins.
- After the cells are classified, unique cells can be imaged with unique colors.
- Unique mRNA profiles can be used in place of unique surface proteins. This method can be used to embed molecular processing data within imaging.
- Water samples can be freeze dried for detection of antibiotics, small molecules, and metals. DNA based neural networks can be used for in field classification of water samples based on contaminants.
- The fundamental question that motivated my research is this – there is a continuous spectrum between matter and life. As we understand the spectrum, we can explain how life arises from a collection of lifeless molecules and engineer forms of matter with desired mixtures of life-like and nonlife-like properties.
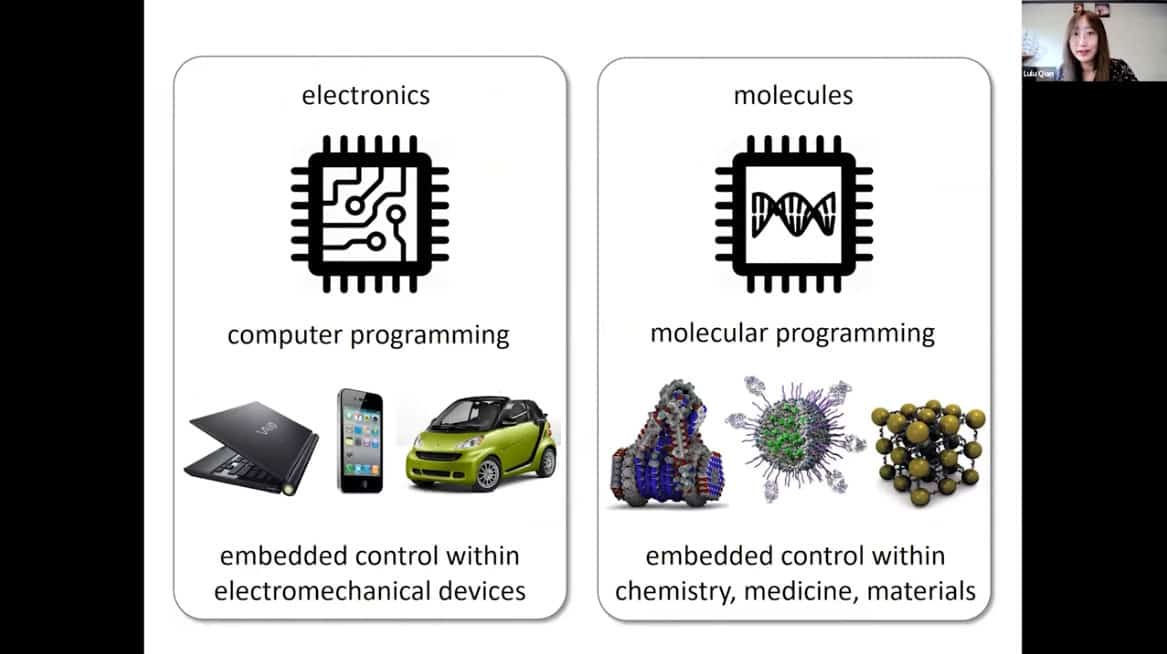
- Similarities – encoded information determines how an entity performs a task. The design space of computer programs is rich and matured. The design space of molecular programs is yet to be explored. However, molecular programs are much more powerful.
- Molecular machines are key to closing the gap. With advances in chemistry the substrate of life has become available for programmable matter.
- Engineering concepts used for computers must be transferred to molecular programming, such as formal programming languages.
- A 30 year challenge is to build a synthetic cell, with a synthetic level of complexity similar to a biological cell.
- By building this system we will be able to learn how to design structural, mechanical, and computational components that are fully composable and programmable. We will learn how to store and consume chemical energy over repeated cycles and demonstrate sophisticated behaviors like learning and self-improvement.
- There are several challenges associated with this goal.
- Learning allows life to be born simple but develop into high complexity. Two general principles need to be investigated – adaptive memories and self-improvement in molecular circuits.
- Adaptive memories – molecules encoding memory can be inhibited or activated
- Self-improvement – input signals trigger response to adjust memories which improves pattern recognition in the future
- Three classes of learning
- Supervised learning – circuits exposed to what they may encounter in the future and desired outcomes Unsupervised learning – circuits exposed to what they may encounter, but not to ideal outcomes Reinforcement learning – exposed to same information as unsupervised learning but then that information is used to take an action, which triggers a reward or punishment
- Weight activation needs to be adjustable for each type of learning
- To achieve this type of learning, the organism must be capable of repeated learning events. The current process uses up the gate molecules which means it cannot be used repeatedly.
- The challenge of learning is closely tied to the challenge of metabolism. In fact, all of these challenges are connected in one way or another.
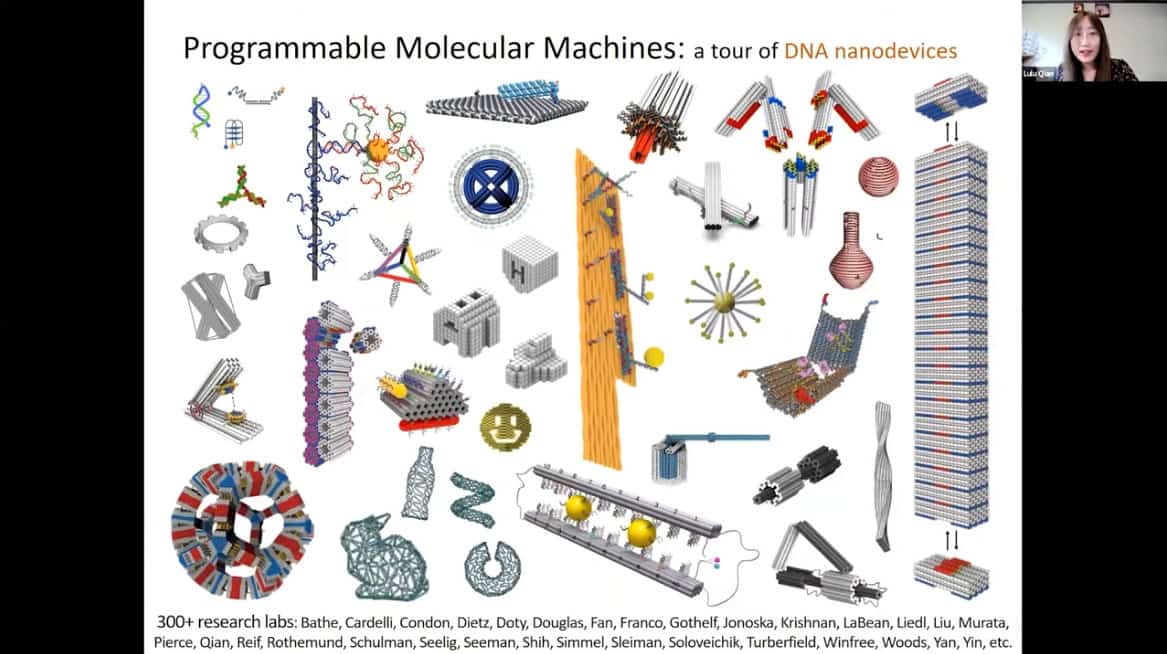
- The first synthetic DNA nanostructure was created in 1983, the first synthetic DNA computing device was created by a computer scientist in 1994. Since then, many more synthetic devices and structures made of DNA and RNA have emerged.
Biomolecules provide the ideal substrate for a new programmable matter, enabling powerful and embedded control within chemistry, medicine, and materials.
End of presentation