Since its invention in 2006 by Paul Rothemund, scaffolded DNA origami has been used to build increasingly complex 2D and 3D structures, including organizing nanoscale functional objects in 3D space. The past few years have seen increasing progress in extending the usefulness of this very versatile method of constructing nanostructures from DNA. Three years ago we noted the report from Hendrik Dietz’s group of improvements in technique that resulted in faster folding, gave higher yields, and provided structures assembled with sub-nanometer positional precision. Higher yields of more precisely assembled DNA nanostructures opened the way to using DNA origami to build more dynamic structures evincing mechanical movements. A year ago we cited the accomplishments of a former postdoctoral fellow in Dietz’s group, Carlos Castro, in achieving well-defined programmed motions with DNA nanostructures, thus beginning to fabricate parts for machine designs based upon the way that macroscopic machines work. We reported additional progress from Castro and his colleagues here and here. A few months later we cited a report from Dietz’s lab on DNA components with complementary shapes that self assemble into nanoscale machinery. Finally, last December we noted the accomplishment of the Dietz group in using DNA nanotechnology to position molecules with atomic precision. A new result from the Dietz group recently published in the open access journal Science Advances and also available from Dietz’s publications page demonstrates (quoting from the abstract) “a nanoscale rotary mechanism that reproduces some of the dynamic properties of biological rotary motors in the absence of an energy source, such as random walks on a circle with dwells at docking sites.” Both this and the previous advance of positioning molecules with atomic precision are described in a press release from the Technical University of Munich “Nanoscale rotor and gripper push DNA origami to new limits“:
Dietz lab’s latest DNA nanomachines demonstrate dynamics and precision
Scientists at the Technical University of Munich (TUM) have built two new nanoscale machines with moving parts, using DNA as a programmable, self-assembling construction material. In the journal Science Advances, they describe a rotor mechanism formed from interlocking 3-D DNA components. Another recent paper, in Nature Nanotechnology, reported a hinged molecular manipulator, also made from DNA. These are just the latest steps in a campaign to transform so-called “DNA origami” into an industrially useful, commercially viable technology.
Inspired by nature’s nanomachines – such as the enzyme ATP synthase and the motor-driven flagella of bacteria – physicists in Prof. Hendrik Dietz’s lab at TUM keep expanding their own design and construction repertoire. They have systematically developed rules and procedures for creating self-assembled DNA origami structures with ever greater flexibility and control. Moving from DNA basepair matching to shape-complementary building techniques – with a variety of interlocking “bricks” – the researchers’ toolkit has advanced steadily in the direction of higher-level programming and modular assembly.
In step with this progress, they have honed methods needed to verify, for example, that a particular soup of nanoparticles really is packed with copies of whatever they designed: whether a switchable gear, an artificial membrane channel, an arbitrarily complicated test object, a “nanobook” that opens and closes, or a robot figure with movable arms.
Two new additions to the zoo
The latest additions to the lab’s zoo of DNA origami objects, two tiny 3-D machines with moving parts, demonstrate new capabilities. (See video at EurekAlert, 1’23”.)
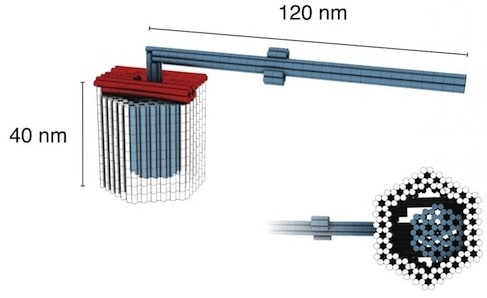
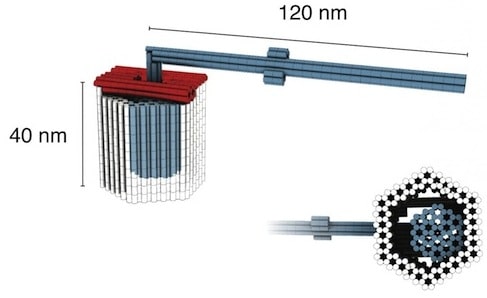
Doctoral candidates Philip Ketterer and Elena Willner collaborated with Dietz in building a rotory [sic] mechanism from three multilayer DNA building blocks: a rotor unit, with a body roughly 32 nanometers long and a longer, lever-like extension; and two clamp elements that “click” together to form an axle bearing. The parts join with a tight fit and leave just 2 nanometers of play around the axle, allowing the rotor to swing but not to wobble. In one variant, the arm will rotate freely between random stopping points; in another, it will dwell in specified positions the researchers call docking sites. To date, this is the most complex rotary structure realized using DNA origami techniques.
To be clear, the rotor has no motor: It is propelled by Brownian motion, random movement of particles in solution. By demonstrating the feasibility of building such a machine, however, the researchers open the way for active devices under chemical or thermal power and control. “It’s like having built an engine,” Dietz says. “Now spark plugs and combustible fuel are the next items on the to-do list.”
In a separate project, Dietz and doctoral candidate Jonas Funke created a hinged machine on a scale suitable for manipulating individual molecules with atomic precision. The angle between the gripper’s two main structural elements can be controlled with DNA helices. Experiments with this DNA origami positioning device showed that it could be capable of precisely placing molecules, adjusting the distance between them in steps as small as the radius of a hydrogen atom. This work is significant in that it bucks a trend in the field toward building larger DNA origami devices without necessarily pushing the limits of precision. In addition, the results hint at one of the ways DNA nanomachines might someday be useful to control chemical reactions.
“On the one hand,” Dietz says, “we now really trust the placement precision within our structures – because we have actually placed two molecules and controlled their distance with atomic precision. On the other hand we have now a prototype rotary mechanism. Based on our measurements, its mobility is such that it could do up to 50,000 rpm if every rotary step it took would go only in one direction. In the next generation of devices, we will use the placement precision to couple chemical reactions to the movements of a rotor. This has the potential to result in a motor. This device, then, could be used for all kinds of purposes, such as actively propelling nanoscale drug-delivery vehicles, pumping and separating molecules across barriers, or packaging molecules into cargo compartments.”
Work with dynamic DNA nanostructures is clearly heading in a very interesting direction. Providing a motor to power a mechanism such as the one demonstrated in this paper, instead of relying on Brownian motion, would be a very important next step. Perhaps a few more generations of improved devices will produce nanomachines that will be able to make or break specific chemical bonds? Increasingly complex systems of such devices could perhaps provide a path to productive nanosystems and eventually to atomically precise manufacturing.
—James Lewis, PhD