Our most recent post cited research that demonstrated programmed complex motions in simple mechanisms fabricated using scaffolded DNA origami. This achievement represents initial success in what appears to be a systematic program to implement macroscale engineering design principles in molecular machinery made possible by structural DNA Nanotechnology. This approach was introduced in a 2013 paper published in ACS Nano implementing a collaboration between a DNA nanotechnology research group and a mechanical engineering and kinematics design innovation and simulation research group. From “DNA Origami Compliant Nanostructures with Tunable Mechanical Properties“:
Abstract: DNA origami enables fabrication of precise nanostructures by programming the self-assembly of DNA. While this approach has been used to make a variety of complex 2D and 3D objects, the mechanical functionality of these structures is limited due to their rigid nature. We explore the fabrication of deformable, or compliant, objects to establish a framework for mechanically functional nanostructures. This compliant design approach is used in macroscopic engineering to make devices including sensors, actuators, and robots. We build compliant nanostructures by utilizing the entropic elasticity of single-stranded DNA (ssDNA) to locally bend bundles of double-stranded DNA into bent geometries whose curvature and mechanical properties can be tuned by controlling the length of ssDNA strands. We demonstrate an ability to achieve a wide range of geometries by adjusting a few strands in the nanostructure design. We further developed a mechanical model to predict both geometry and mechanical properties of our compliant nanostructures that agrees well with experiments. Our results provide a basis for the design of mechanically functional DNA origami devices and materials.
The authors note that controllable mechanical behavior is achieved by using multiple components with the desired mechanical stiffness. Scaffolded DNA origami can be used to make very stiff structures of bundles of double stranded-DNA (dsDNA), so the range of values of mechanical stiffness available for designs varies over a thousand-fold range from single-stranded DNA to bundles of dsDNA. In macroscopic systems, joints integrate flexible elements with constrained motion, but flexibility in nanoscale systems leads to random thermal motions. In other microscopic systems, compliant mechanism design has been used to achieve controlled mechanical and dynamic behavior with stiff components. To establish a foundation for DNA origami compliant mechanisms, this paper presents “a deformable (compliant) DNA origami nanostructure with controllable geometry and mechanical behavior.”
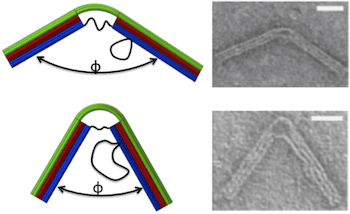
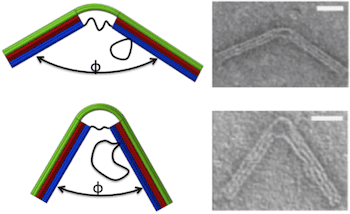
The nanostructure was designed to behave like a hinge with a torsional spring. An 18-helix bundle was organized into three layers of six each. The two ends contain all 18 helices and are therefore stiff enough to be integrated into a larger structure. The central portion comprises the top layer of six helices, which is continuous with the top layer of the two ends, and six ssDNA connections between the two ends of the bottom layer, which function as entropic springs causing the top layer to bend. The length of the ssDNA springs determines the bending force on the joint. Scaffold loops on one side of the hinge provided a reservoir of ssDNA that could be used to add to the length of the springs.
Hinge geometries were confirmed by transmission electron microscopy, yielding Gaussian distributions of joint angles. As the length of the springs increased from 11 to 74 bases, the joint angle corresponding to the peak of the Gaussian increased from 55 ° to 130 °. These experimental results provided a good fit to an analytical model based on a wormlike chain model for the ssDNA springs and a beam model for the 6-layer helix bundle. Further, the Gaussian distribution of angles was wider for longer springs, due to increased thermal fluctuations, consistent with torsional stiffness decreasing as joint angle increases. Thus, the stiffness of the compliant joint is tunable. Calculated torsional stiffnesses varied from 107 pN-nm/rad for the longest spring to 367 pN-nm/rad for the shortest, similar to those of actin-binding proteins.
The authors point out that for these results they have used only one of three design parameters available to them to tune joint stiffness. By also varying the length of the stiff component (the six-dsDNA helices of the top layer) and the stiffness of the stiff component (the number of dsDNA helices), both the bending stiffness and joint angle could be independently designed.
The authors conclude:
Our approach establishes a foundation to design and fabricate DNA-based devices with mechanically functional components such as springs, joints, or actuators. Ultimately, compliant components like the ones presented here can form the basis of compliant mechanisms that can be applied as nanomechanical devices with controllable motion.
The results this collaboration presented recently demonstrate the utility of this foundation presented just a litte more than a year earlier. It will be interesting to see to what extent macroscale engineering design principles can be implemented in DNA molecular machine systems.
—James Lewis, PhD