The idea of using a DNA framework as a “nanobreadboard” to prototype various nanoscale circuits and device arrays goes back at least to Paul Rothemund’s 2006 invention of scaffolded DNA origami technology. The idea played a central role in the development of the concept of modular molecular composite nanosystems formulated as part of the 2007 Foresight and Battelle Technology Roadmap for Productive Nanosystems. Earlier this year researchers at Boise State University in Idaho published an open access article in the journal ACS Photonics “Excitonic AND Logic Gates on DNA Brick Nanobreadboards“. However, instead of scaffolded DNA origami, these researchers built their nanobreadboards using an alternate form of structural DNA nanotechnology (see “Arbitrarily complex 3D DNA nanostructures built from DNA bricks“) that has been extended to fabricate micrometer-scale structures that offer unique opportunities as molecular “breadboards”.
To make single-molecule optical devices for computing and other applications, which exploit the interactions between light and matter at much smaller length scales than the free-space wavelength of light, these molecular chromophores must be precisely arranged to enhance non-radiative dipole-dipole coupling between neighboring chromophores. subnanometer resolution. This coupling facilitates an energy transfer process (FRET – Förster Resonance Energy Transfer) that occurs over a distance that is typically 5 nm or less. Molecular orientation also plays a role. Previous studies of multi-chromophore excitonic circuits have positioned the chromophores using single DNA duplexes or multiarm DNA junctions, or DNA origami. With DNA origami, however, it is only practical to conjugate chromophores to selected staple strands; the long scaffold strand can only play a role in the overall structure since conjugation to the scaffold strand is impractical. This restricted role of the scaffold strand limits the feasibility of using DNA origami for the rapid prototyping of excitonic circuits that is essential to achieve complex functionality.
DNA brick-based nanobreadboards, however, can be assembled by selecting components from a master DNA library. Different complex shapes can be obtained merely by selecting different subsets of the master library, while DNA origami would require re-designing all the staple strands for each new breadboard. The authors also claim that because of the absence of a scaffold strand and because any DNA brick can carry covalently attached chromophores, packing density and position control is twice what it would be with DNA origami. Capitalizing on these advantages:
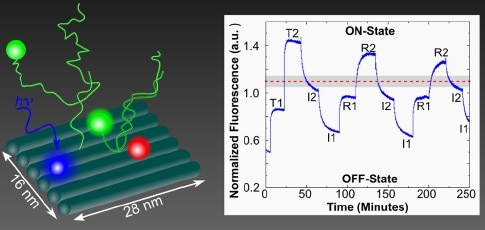
… In this work, we show (1) four-chromophore systems with two dynamically controlled inputs permitting bilevel switches that can be coupled to demonstrate fully excitonic AND logic, and (2) dynamic excitonic switching and logic operations controlled by isothermal DNA reactions in stoichiometric quantities. …
Although two AND gates are described (each using four chromophores in different spatial arrangements), the authors claim that any and all logic gate operations can be implemented. Further, the coupling of multiple excitonic devices on a nanobreadboard promotes achieving greater circuit complexity. In each design, two chromophores are permanently attached to the nanobreadboard, while the other two are added to or removed from DNA tethers by strand displacement. When both of these are attached to the nanobreadboard, they complete a FRET transmission, and the gates are in the ON-state. Both AND gate designs showed successful switching, and the procedure clearly leaves room for further optimization. This paper provides another indication that we are still in early days for determining the functional potential of DNA nanotechnology and modular molecular composite nanosystems.
—James Lewis, PhD
