Four years ago we cited a report by a German research group of a single molecule cut and paste technology to assemble molecular building blocks on a DNA scaffold. The advance was noteworthy because it combined self-assembly of atomically precise components with the ability to use a manipulator (an atomic force microscope) to place those components at arbitrary positions in a larger structure, analogous to the way in which we use our hands to assemble parts macroscopically. These researchers have extended this technology to arrange single protein molecules. A hat tip to ScienceDaily.com and KurzweilAI.net for pointing to this press release from Ludwig-Maximilians University in Munich “All systems go at the biofactory“:
In order to assemble novel biomolecular machines, individual protein molecules must be installed at their site of operation with nanometer precision. LMU researchers have now found a way to do just that. Green light on protein assembly!
The finely honed tip of the atomic force microscope (AFM) allows one to pick up single biomolecules and deposit them elsewhere with nanometer accuracy. The technique is referred to as Single-Molecule Cut & Paste (SMC&P), and was developed by the research group led by LMU physicist Professor Hermann Gaub. In its initial form, it was only applicable to DNA molecules. However, the molecular machines responsible for many of the biochemical processes in cells consist of proteins, and the controlled assembly of such devices is one of the major goals of nanotechnology. A practical method for doing so would not only provide novel insights into the workings of living cells, but would also furnish a way to develop, construct and utilize designer nanomachines.
In a major step towards this goal, the LMU team has modified the method to allow them to take proteins from a storage site and place them at defined locations within a construction area with nanometer precision. “In liquid medium at room temperature, the “weather conditions” at the nanoscale are comparable to those in a hurricane,” says Mathias Strackharn, first author of the new study. Hence, the molecules being manipulated must be firmly attached to the tip of the AFM and held securely in place in the construction area.
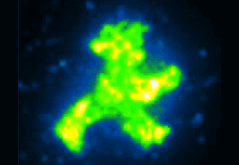
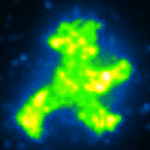
The forces that tether the proteins during transport and assembly must also be weak enough not to cause damage, and must be tightly controlled. To achieve these two goals, the researchers used a combination of antibodies, DNA-binding “zinc-finger” proteins, and DNA anchors. “We demonstrated the method’s feasibility by bringing hundreds of fluorescent GFP molecules together to form a little green man, like the traffic-light figure that signals to pedestrians to cross the road, but only some micrometers high,” Strackharn explains.
With this technique, functional aspects of complex protein machines – such as how combinations of different enzymes interact, and how close together they must be to perform coupled reactions – can be tested directly. A further goal is to develop artificial multimolecular assemblies modeled on natural “cellulosomes”, which could be used to convert plant biomass into biofuels. Strackharn points out the implications: “If we can efficiently build mimics of these ‘enzymatic assembly lines’ by bringing individual proteins together, we could perhaps make a significant contribution to the exploitation of sustainable energy sources.” [abstract of research paper]
The precision achieved in this preliminary report of single-molecule cut-and-paste with proteins is about 10 nm. This is about 100-fold less precise than atomic precision. However, the AFM tip can be moved with atomic precision (0.1 nm). The lower precision seen in this preliminary demonstration is due to the long spacer used to attach the anchor DNA to the surface, so the precision can presumably be improved in future work as it becomes more important.
To use the AFM to build a structure by precisely placing GFP molecules, the Green Fluorescent Protein was fused to a zinc finger protein that binds a specific fragment of double strand DNA. This DNA fragment has a single strand overhang that binds to a single strand anchor DNA on the surface in what the authors refer to as the “unzip” geometry so that the DNA binding can be pulled apart one base pair at a time. The GFP is also fused to a short peptide fragment that will bind to an antibody fragment that is attached to the AFM tip.
The combination of DNA base pair interactions and peptide-antibody interactions allows controlled transfer of the green fluorescent protein-DNA complex from the depot to the AFM tip because only a force of 25 pN is required to separate the complex from the depot one base pair at a time, but a force of 40 pN binds the complex to the AFM tip. Lowering the AFM tip at the target position binds to the anchor DNA at the target, but because the DNA strand at the target position is in the “shear” geometry, this new DNA complex cannot be ruptured by a force of less than 60 pN, so the AFM tip can be retracted leaving the GFP complex in the new position.
Assembling the micrometer-size image of the “traffic man” from individual GFP molecules required 900 steps. Two “traffic man” icons were assembled: one using a red dye coupled to the DNA component of the complex that was assembled in the shape of the sign that means “Don’t walk,” and one using the green fluorescence of the GFP itself in the shape of the sign that means “Walk”. The fact the GFP still fluoresces shows that the forces used during the process are gentle enough to preserve the function of the protein.
This work is an important step in the biology-based folding polymer path toward designing and building complex molecular machine systems (see the Technology Roadmap for Productive Nanosystems for discussions of various paths). With the huge array of molecular machines that biology provides it should be possible to explore the capabilities of some very sophisticated systems. It is not yet clear just how precise this cut and paste system can be made, so it is premature to compare it with other proposed systems of assembling designed arrays of proteins: using DNA origami or DNA tiles as scaffolds.
—James Lewis, PhD