A select set of videos from the 2013 Foresight Technical Conference: Illuminating Atomic Precision, held January 11-13, 2013 in Palo Alto, have been made available on vimeo. Videos have been posted of those presentations for which the speakers have consented. Other presentations contained confidential information and will not be posted.
The second speaker at the Computation and Molecular Nanotechnolgies session, Art Olson, presented “New Methods of Exploring, Analyzing, and Predicting Molecular Interactions” . https://vimeo.com/63008844 – video length 46:17. Prof. Olson began with three simple points on interacting with and understanding the nanoscale world: (1) human interaction—how we understand something that we can’t see directly; (2) how we integrate data from lots of different sources to form a picture of what is happening at the nanoscale; (3) how we develop software tools to move forward in these areas.
Olson recommended that we should use all of our senses as much as we can because we learn in different ways; we see in different ways; we understand in different ways. Similarly, there is no one data source that gives us a complete picture of the molecular world, so we have to synthesize across scales, and we have to synthesize across methods. With software, collaboration is important, because if we can bridge disciplines rather than reinvent what already exists, we can move a lot faster.
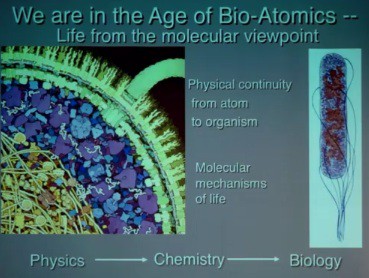
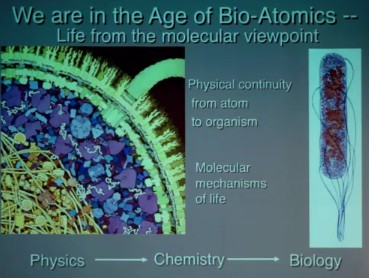
As a molecular biologist studying biological systems, Olson emphasized the physical continuity from atom to organism. No mysterious forces are introduced between atom and organism, but the molecular mechanisms of life introduce complexity. Physics provides the fundamental laws governing the interactions of particles at all levels, but complexity derived from combinatorics—how different atoms are combined—ensures that chemistry cannot be predicted from physics. One estimate of the number of small organic compounds that could be made from the atoms present in the human body is 1060. So chemical space is finite but huge, as is biology space. The complexity of biology space includes happenstance—it deals with how matter interacts with its environment. Since the complexity of biology is huge compared to the complexity of chemistry, we cannot predict biology from chemistry alone. We have to understand how evolution factors into this complexity, Olson explained.
Olson noted the availability of various techniques for determining the structures of life that span time and spatial scales from atoms to organisms, from x-ray crystallography and scanning probe microscopy to light microscopy. For the area in the middle—the mesoscale—no one technique can provide the picture alone. We need to synthesize from a variety of techniques.
Olson cited the memoirs of both Watson and Crick as to how important their iconic physical model of DNA was to their interpreting the x-ray fiber diffraction data that Rosalind Franklin had obtained. Olson compared this model to an analog computer—it was a framework on which to play out ideas. An interesting point is that they do not mention the model in their 1953 paper on the DNA double helix, implying to Olson that as scientists, we expound our ideas, but we do not talk about how we came to our ideas. Models can play an important part in the early stages of how we think about the molecular world.
Citing the well-known model of a kinesin molecule walking along a microtubule in a stochastic process that nevertheless moves in only one direction, Olson pointed to a paper from others describing an in vitro system comprising four types of molecules: the microtubule, kinesin, streptavidin to which multiple kinesins can be bound to make them multivalent, and a crowding agent (polyethylene glycol). The crowding agent drives these microtubules decorated with kinesins into long fibers. The fibers have a polarity according to whether the kinesins are walking up the fiber or down the fiber. For reasons not understood, the bundles elongate until, at a certain length, they buckle. With the addition of energy in the form of ATP, a large scale motion appears. Placed in a bubble large enough for the buckling to appear, a dynamic structure appears on the surface. These bubbles are about the size of cells (microscopic rather than nanoscopic) move around like cells. If these large bubbles are placed on surfaces, they tend to coordinate with each other. All of this emergent behavior arises from a system with just four molecular components, Olson emphasized. Not only is structure important, but dynamics is extremely important in understanding what is going on.
Turning next to the human-computer interface, Olson emphasized that (1) biological structures are complex, (2) spatial relationships and reasoning are difficult, (3) mouse and keyboard are limiting, and (4) images and computer graphics may not convey the whole picture. Human-computer interaction has changed dramatically with tablets, motion detectors, video everywhere, and 3D-printing (making macroscopic models because they have perceptual advantages).
Quoting artificial intelligence pioneer Marvin Minsky “If you ‘understand’ something in only one way, the you don’t ‘really’ understand it at all”, Olson described how he found that to be true with protein structure. Physical models promote perceptual integration (vision and tactile). It is also a rich cognitive substrate, like Watson and Crick’s DNA double helix model. Movement is intuitively intelligible so that instead of thinking about moving it, you can think about the implications of moving it. A physical model is also an analogical and metaphorical medium for exploration, promoting serious play as an important part of the creative process. It also promotes social cognition by sharing an object. Using a physical model of the polio virus, Prof. Olson demonstrated that it can assemble through random motion by shaking it.
Olson presented augmented reality as one way to combine the physical model with the computer model. He demonstrated a physical model representing protein folding using magnets to represent hydrogen bond donors and acceptors. the model could be physically manipulated to make alpha helices, beta sheets, etc. The physical model can act as a computer interface, manipulating the computer model by manipulating the physical model.
Next focusing on the kinds of data that are revealed by these tools, Olson showed a water color painting by his collaborator David Goodsell. These paintings changed how molecular and cell biologists see the cell by making visible the concentration, variety, and compartmentalization of proteins. The major work, Olson explained, is accumulating all the needed data—proteomics, crystal structures, etc.—to represent data that we cannot image directly.
Graham Johnson came to Olson’s lab as a graduate student to do with 3D models what Goodsell did with his water colors. This is essentially a problem of packing various shapes into a given volume. He and Ludovic Autin devised a grid-baed approach in which you take data from, for example, electron tomography, which gives you the shapes of all of the organelles, you defined the interior and exterior surfaces, you enter the data for what is inside the organelle, what is on the surface, and you use a variety of packing algorithms to generate a packing of all of the shapes into the surfaces and volumes.
Finally, Prof. Olson noted that his group were pioneers in developing molecular modeling and molecular visualization software for the past 30 years, inventing solutions for one domain. Eventually they wre overwhelmed by the advances of the entertainment industry and the gaming industry in interactions, rendering, modeling, etc. because those industries had spent billions of dollars on large packages of sophisticated software. Olson’s group wanted to take advantage of these packages, which could do, for example, fantastically fast collision detection, but could only treat molecules as dead geometry.
By happenstance, thanks to Michel Sanner recommending Python for the Olson group’s development efforts 15 years ago, all of their development has been done in Python. It turns out that all of the high-end packages developed for Hollywood, etc., have embedded Python interpreters. Therefore, the molecular modeling components from the Olson group software can be taken out, matched with a thin adapter, and plugged into any of the high-end computer graphic packages. Thus any domain-specific modeling package can be brought into Maya, or Cinema 4D, or the open source Blender. Now they have a workflow that goes from crystal structures and proteomics data all the way to the full model of, for example, an HIV virus particle or a synaptic vesicle and its associated cytoplasmic proteins. Multiple models incorporating multiple parameters can be generated automatically, and then run to see which ones give behavior compatible with observation.
Prof. Olsen made the point that it is important for scientists to communicate their results to the world. The future of science and technology lies in the hands of future scientists and technologists. To educate them, we need tools to get them excited about things they have no physical experience with. Olsen has founded a small company named Science Within Reach to provide some of these tools to high school and middle school students.
—James Lewis, PhD