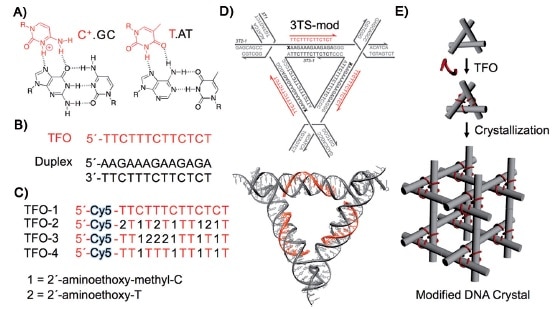
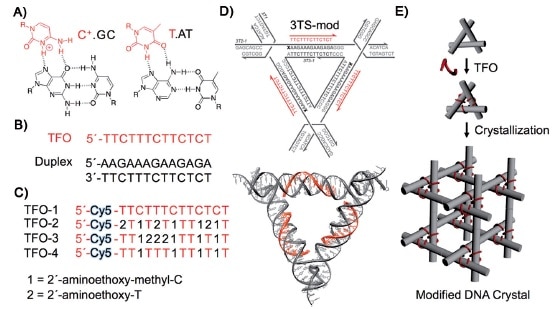
Our previous post focused on the production of high quality macroscopic DNA crystals containing fairly large (on a molecular scale) cavities. This post deals with the challenge of precisely filling those cavities with guest molecules or nanoparticles. In 2014 Seeman and his collaborators reported using triplex forming oligonucleotides to programmably position guest components on the double-helical edges of the tensegrity triangles comprising the crystal: “Functionalizing Designer DNA Crystals with a Triple-Helical Veneer” [OPEN ACCESS]. Citing their earlier work reporting crystals with cavities exceeding 1000 nm3, the authors propose introducing guest molecules into these cavities by targeting a DNA sequence within the tile comprising the crystal, using triplex-forming oligonucleotides that bind in a sequence specific fashion to the major groove of the DNA double helix by forming base triplets. Because triplex formation requires a lower pH, some triplex forming oligonucleotides incorporated triplex stabilizing nucleosides in place of the usual DNA nucleosides C and T. A cyanine dye molecule was attached o he 5`-terminus of each triplex forming oligonucleotide (TFO) to facilitate characterization of the product and to serve as a test guest molecule to be incorporated into the crystal.
TFOs were shown to bind to the tensegrity triangle tiles as expected. Binding of the TFOs did not affect the formation of crystals from the tiles. Fluorescence of the crystals clearly showed that the dye had been incorporated. Several of the crystals were analyzed by X-ray diffraction, yielding the same results as the previous work.
The crystals produced in this study were about 100µm on a side, containing an estimated 1012 unit cells. The same TFO is targeted to each helix within the crystal, which means each guest is positioned with sub-nanometer precision. Three guest components are housed within each of the crystal’s 366 nm3 rhombohedral cavities. The authors note that each guest component is separated by ~10.5 nm along the helix axis between tiles, and by 5.8 nm through 3D space within the tile. The authors note that the diffraction resolution of these crystals is is relatively low, 0.6 nm, but that this might be improved to 0.4 nm by targeting guests to two helical turns per edge instead of three, thus producing crystals with less hollow space that therefore do not deform as easily. The authors remain focused on the goal of using these crystals as hosts to alleviate the difficulties of macromolecular crystallization for X-ray diffraction analysis.
—James Lewis, PhD