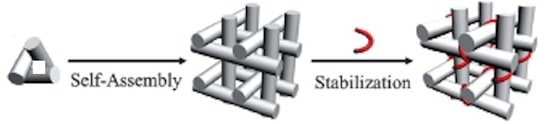
To self-assemble macroscopic, porous DNA crystals suitable for use as structural scaffolds or molecular sieves, it was first necessary to show that macroscopic crystals could be self-assembled from atomically precise DNA nanostructures, and then to show that triple helix forming oligo nucleotides could target cargo molecules to each cavity in the crystal with sub-nanometer precision. A paper published last year from Prof. Chengde Mao of Purdue University and Prof. Nadrian C Seeman of New York University, and their collaborators tackles and resolves conflicting requirements for successful self assembly. The component nanostructures must attach to each other using interactions that are weak enough that a building block in an incorrect site can detach, but strong enough that the final structure is stable. They report success using another molecule that binds to the cohesive sites and stabilizes the interactions among the subunits: “Post-Assembly Stabilization of Rationally Designed DNA Crystals” (abstract, full text behind pay wall).
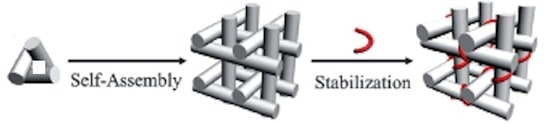
The authors explain that the triangular DNA motifs that they engineered to assemble into macroscopic 3D crystals bind weakly to adjacent motifs through a pair of two-nucleotide single-stranded overhangs (sticky ends). With such weak interactions, the crystals are stable only in high salt solutions (> 1.2M (NH4)2SO4), which however, greatly limit the applications for which they can be used. Their solution is to strength the inter-triangle interactions after self-assembly by adding a triplex-forming oligonucleotide at the inter-triangle cohesion region to enhance the interactions of the tensegrity triangle subunits.
To accomplish this post self-assembly stabilization the tensegrity triangle subunit motif was modified such that the DNA duplex region that connects two adjacent triangle motifs contains only purines on one strand and only pyrimidines on the other. At an acidic pH of 5.6 this DNA duplex region can bind to a pyrimidine-only strand to form a triplex, which stabilizes the sticky-end cohesion, and thus the DNA crystal so that it is stable in low ionic strength solutions, such as 0.02M (NH4)2SO4. The authors demonstrate that without triplex formation, the crystals formed at high ionic strength (high salt) (1.7M) disintegrate when exposed to even a small decrease in ionic strength (1.2M). With the triplex, the crystals were stable at the lower ionic strength, and even at very low ionic strength (0.02M), although they too disintegrated when incubated against water without any salt. Control experiments adding single strands not capable of triplex formation instead of the triplex forming strand, or adding the triplex forming strand at pH 8.0 instead of pH 5.6, had no effect on stability of the crystals because no triplex formed.
Other experiments demonstrated that crystals stabilized by triplex formation had about 50% of the sticky end cohesions stabilized by triplex formation, so that it is not necessary to stabilize each cohesive end in the crystal to obtain stabilized DNA crystals. The authors point out that further methods of stabilization, such as using modified nucleosides to facilitate triplex formation at neutral pH, or using sequence specific protein binding to the sticky-end cohesion, could be investigated. Stabilization of crystals under more physiological conditions would permit applications with guest molecules, like enzymes or nanoparticles, that are also more active under physiological conditions.
In theory,the problem addressed by this paper could be solved by either chemical or enzymatic ligation of the cohesive ends after crystal formation, thus substituting strong covalent bonds for weak interactions; in practice, both enzymatic and chemical ligation of large DNA nanostructures have proven problematic (Nadrian C. Seeman, 2015). For the mostly biotech applications suggested by the authors, requiring macroscopic DNA crystals, the approach described here may be the best current bet. However, as a step toward productive nanosystems leading eventually to atomically precise manufacturing, smaller scaffolds containing hundreds or thousands instead of trillions of subunits might suffice. The best approach to make scaffolds in this size range may be an open question still.
—James Lewis, PhD