In writing this blog I occasionally find that I missed work that is an important part of a developing story. Over the years, DNA nanotechnology has been one of the most promising paths from current nanoscience and nanotechnology to the ultimate goal of general-purpose high-throughput atomically precise manufacturing. Most of the recent progress we have reported on DNA nanotechnology has been based upon the scaffolded DNA origami technique or, to a lesser extent, the DNA bricks technique. The original introduction to DNA nanotechnology came in a 1987 paper by DNA nanotechnology pioneer Nadrian Seeman and his chemist collaborator Bruce Robinson “The design of a biochip: a self-assembling molecular-scale memory device” (full text behind a pay wall), which proposed a self-assembling DNA scaffold that could serve as a framework for a molecular wire and switch. This proposal was based on Seeman’s earlier (1985) suggestion that ligation of DNA branched junction building blocks could lead to a periodic array, analogous to the crystallization of molecular systems.
Three papers by Seeman and his collaborators published the last several years (2009, 2014, 2015) highlight progress toward Seeman’s original vision of a practical macroscopic DNA crystal array with cavities that are large on the molecular scale and could be used to precisely order molecular components in three dimensions. This post considers the earliest of these. The first step from concept to reality is explained by the title of the article “From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal“. The full text is behind a pay wall, but a free PubMed Central version is available.
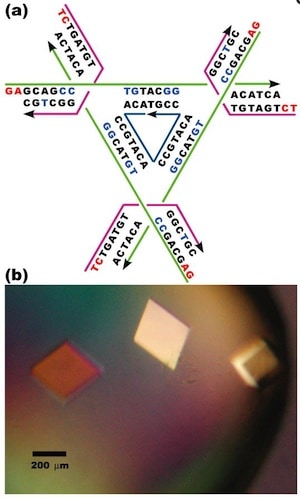
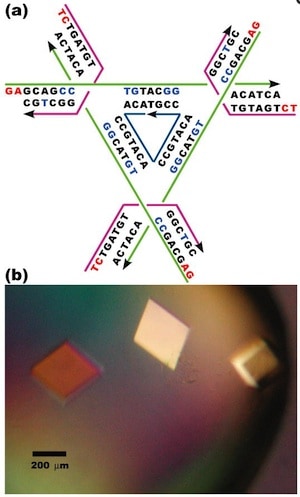
The authors note that producing a macroscopic 3D periodic array of precisely arranged molecular components should be possible with appropriate branched DNA motifs with ‘sticky’ tails. “It is essential that the directions of propagation associated with the sticky ends do not share the same plane, but extend to form a 3D arrangement of matter.” Based on a ‘DNA tensegrity triangle’ motif that they had reported in 2004, they report here the construction of ~250-µm-sized DNA crystals, five orders of magnitude larger than the nanometer scale of molecular components.
The authors explain that the tensegrity triangle motif is made rigid because it is constructed from three DNA helices connected pairwise by three four-arm branched junctions to produce a stiff alternating over-and-under motif. The axes of the three helices are directed along linearly independent vectors so they are not coplanar. The design the authors chose to work with has an edge length of 21 nucleotide pairs. By extending the three helices with single-stranded ends that can be used for self-assembly, the helices can link to six other molecules in six different directions, producing macroscopic (> 0.25 mm) rhombohedral crystals. Iodinated heavy atom variants of the crystals were made to facilitate X-ray diffraction analysis of the crystal structure, allowing the authors to report the crystal structure to a resolution of 0.40 nm—not enough to discern atomic detail, but enough to discern DNA double helical structure. The authors report that the DNA is mostly in B form, as expected, but certain specific nucleotides in the triangle are closer to the A form. They also report that the major and minor grooves of the helix can be discerned.
The authors further report that the rhombohedral lattice yields a unit cell with a side of 6.922 nm and an angle between the edges of 101.44°, which according to the rhombohedron calculator yields a surface area of 281.773 nm2 and a volume of 308.708 nm3. The authors state that the volume of the rhombohedral cavity is ~103 nm3, and its open cross section is ~23 nm2.
The authors constructed eight other rhombohedral crystals using variations on the first triangle sequence, with edge lengths varying from 21 to 42 nucleotide pairs. These produced rhombohedral cell dimensions varying from 6.80 nm and 102.6° to 13.49 nm and 117.3°. The crystals were not as rigid and precise as the first one reported above, with structure determined to a resolution varying from 0.50 nm to 1.40 nm. The cross section of the internal cavity varied from 23 to 123 nm2, and the volume of the internal cavity varied from 101 to 1104 nm3. As expected, the resolution of the crystals decreases (that is, the length resolved increases) as the edge length increases because of stick-like lattices have more hollow space.
Having established that they can make well-ordered, macroscopic DNA crystals, the next question Seeman and his collaborators tackled, was how to address specific locations on such arrays.
—James Lewis, PhD