Yet another milestone along the protein design molecular engineering path to advanced nanotechnology has been reached, thanks to the efforts of the laboratory of David Baker, one of the 2004 winners of the Foresight Feynman Prize in Nanotechnology for Theoretical work. From KurzweilAI “How to design proteins from scratch“:
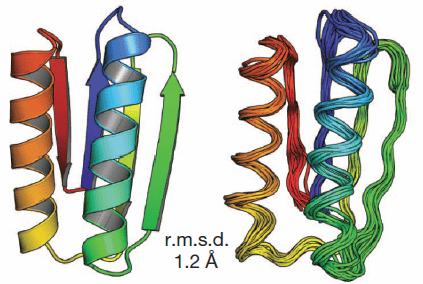
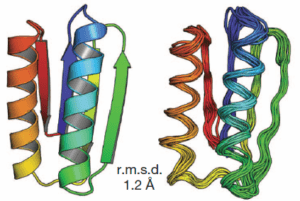
… By following a set of rules, they designed five proteins from scratch that fold reliably into predicted conformations. In a blind test, the team showed that the synthesized proteins closely match the predicted structures.
“What you have now is a flexible set of building blocks for nanoscale assembly,” says Jeremy England, a molecular biophysicist at the Massachusetts Institute of Technology in Cambridge, who was not involved in the work. …
More detail is given in a commentary in Nature, the journal in which the research was published “Proteins made to order: Researchers design proteins from scratch with predictable structures“:.
… Baker’s proteins are in a sense “platonic ideals”, he says: simple backbone constructs with every amino acid optimized to fold into the prescribed, stable structure. In this way they differ from natural proteins, whose folded structures represent a compromise between the competing requirements of optimum folding and biological function, leading to “frustrated” parts of the sequence that may be essential for function but are destabilizing to the fold. As evidence of their stability, the designed proteins melt at about 100 °C, Koga says, compared to 40–50 °C for a natural protein. …
The best summary of the significance of this result is given by the authors in the conclusion of their paper (reference numbers omitted from quotation):
The design principles and methodology we have described should allow the ready design of a wide range of robust and stable protein building blocks for the next generation of engineered functional proteins. Almost all protein design and engineering efforts so far have repurposed naturally occurring proteins that evolved for some other, often unrelated, function. It should now become possible to custom-design protein scaffolds ideal for the desired function, and to build larger assemblies and materials from robust ideal building blocks.
From the standpoint of advanced nanotechnology/molecular manufacturing, the “build larger assemblies” part is especially interesting. The abstract of the research is available on the journal web site, and the authors have made a full text PDF available on the Baker lab web site.
—James Lewis, PhD