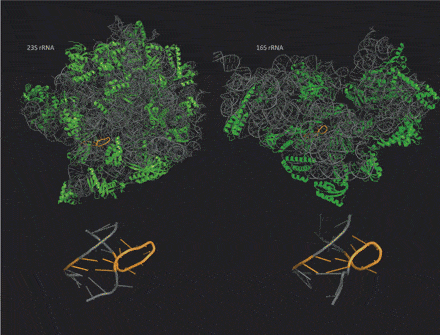
As we pointed out recently, a unique advantage of RNA nanotechnology compared to DNA nanotechnology is that the more complex rules of base pairing involved in RNA folding allow the formation of a variety of compact, complex three-dimensional shapes. Although one principal function of RNA molecules in cells is as messenger RNA, carrying a copy of the information in a DNA gene to the ribosomes where that message is translated to make a protein, a large number of other RNA molecules, including those comprising the ribosomes themselves, have complex three-dimensional shapes and embody various functional properties dependent on those shapes, as do proteins.
Taking advantage of the rapid increase in available high resolution, three-dimensional structures for various non-coding RNA molecules, a new computational method has uncovered many new RNA structural motifs, revealing the tool kit of RNA nanotechnology to be even more diverse than thought. A hat tip to Science Daily for reprinting this news release from the University of Central Florida “Computer Sleuthing Helps Unravel RNA’s Role in Cellular Function“:
… University of Central Florida Engineering Assistant Professor Shaojie Zhang used a complex computer program to analyze RNA motifs – the subunits that make up RNA (ribonucleic acid). …
The units that make up RNA fold like a long accordion and vary in structure. Many have been identified in the past, but finding a quick automatic way to determine patterns in the varying types of units has been elusive until now.
“We have discovered many new RNA structural motifs using our new computational method,” Zhang said. “This breakthrough can largely increase our current knowledge of RNA structural motifs. And newly discovered motifs may also help us develop possible treatment of certain diseases.”Zhang’s work is this month’s cover story in Nucleic Acids Research [abstract, Open Access Full Text], an academic journal.
Using computers, Zhang and his team have been able to view these RNA accordion-like structures and how they fold in a 3-D scale. The program can quickly go through many RNA samples and discover units that are distinct and form patterns. That information gives researchers clues about their function. …
The newly identified structural motifs contain variations in base-pairing rules. As the authors conclude:
These new motifs may lead to the discovery of unknown structure–function relationships and define new building blocks for the RNA architecture, significantly improving our understanding of the RNA structural motifs. …
The next test will be to see if these new insights into RNA structure will enable the design of new RNA machines with novel functions, and eventually artificial RNA molecular machines.
—James Lewis