A study of a biological molecular machine has shown that the machine functions most effectively when it uses chemical bonds just barely strong enough to survive the power stroke of the machine.
A study of a biological molecular machine has shown that the machine functions most effectively when it uses chemical bonds just barely strong enough to survive the power stroke of the machine.

A set of 32-nucleotide single strand DNA bricks was designed so that each can interact independently with four other DNA bricks so that sets of hundreds of bricks can self-assemble into arbitrarily complex 25-nm 3D shapes, each comprising 1000 8-base pair volume elements.
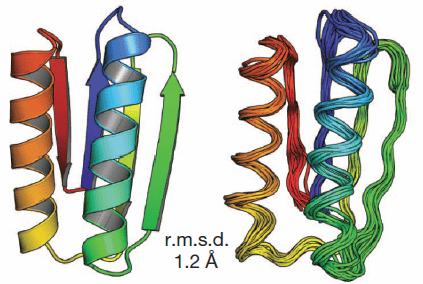
Five proteins were designed from scratch and found to fold into stable proteins as designed, proving the ability to provide ideal, robust building blocks for artificial protein structures.

A single-electron spin qubit on a phosphorous atom in a conventional silicon computer chip has been coherently manipulated, demonstrating the application of single atom nanotechnology to the development of a scalable platform for a quantum computer.

One possible pathway from current technology to advanced nanotechnology that will comprise atomically precise manufacturing implemented by atomically precise machinery is through adaptation and extension of the complex molecular machine systems evolved by biology. Synthetic biology, which engineers new biological systems and function not evolved in nature, is an intermediate stage along this path. An… Continue reading More complex circuits for synthetic biology lead toward engineered cells
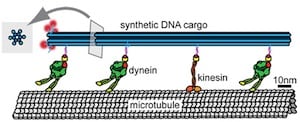
Two types of biological molecular motors that run in opposite directions along a protein track can be used in different arrangements to either move a complex DNA cargo along the track or engage in a tug-of-war.
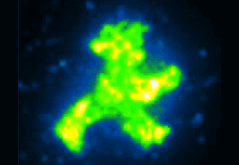
A “cut and paste” method uses an atomic force microscope to assemble protein and DNA molecules to form arbitrarily complex patterns on a surface. Developing this approach to form enzymatic assembly lines could be a path toward a general purpose nanofactory.
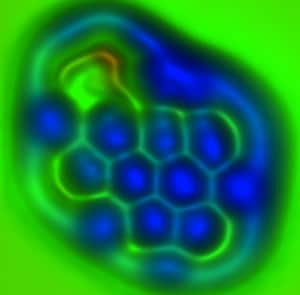
Noncontact atomic force microscopy using a tip functionalized with a single molecule provides highly precise measurement of individual chemical bond lengths and bond orders (roughly, bond strength).
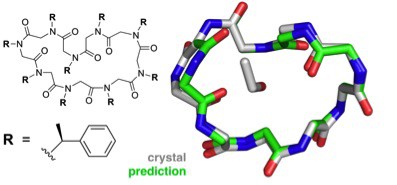
A combination of theoretical and experimental work on peptoids, synthetic analogs of proteins, points to the ability to design peptoids with desired structures and functions.
Researchers have configured a 3D printer as an inexpensive, automated discovery platform for synthetic chemistry. A road to more complex molecular building blocks for nanotechnology?