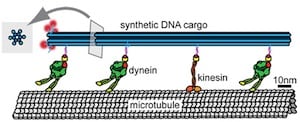
Among the recommendations of the 2007 Technology Roadmap for Productive Nanosystems is the development of modular molecular composite nanosystems (MMCNs), such as systems in which million-atom-scale DNA frameworks are used to organize various functional molecular components in ways to accomplish specific functions, eventually including atomically precise manufacturing. A step in this direction was taken by Harvard University scientists who used a DNA origami framework as a chasis on which to assemble and test the biological molecular motors that maintain subcellular organization in eukaryotic cells through the organized transport of various molecular cargos. In cells these molecular motors dynein and kinesin transport cargos in opposite directions along a hollow 25-nm-diameter protein track—the microtubule component of the cytoskeleton. In this work, the molecular motors carried a DNA chasis cargo along microtubules for a few tens of micrometers—comparable to the length of a eukaryotic cell. “Tug-of-War in Motor Protein Ensembles Revealed with a Programmable DNA Origami Scaffold” was published online in Science last week [abstract, PDF made available by corresponding author].
The DNA chasis comprised a 12-helix bundle with six DNA double helices on the inside and six on outside of the bundle. A total of 90 unique DNA handles on the outer helices of the chasis, and complementary DNA handles on the cargo-binding domains of the dynein and kinesin motor molecules, made it possible to bind specific motor molecules to specific spots on the DNA chasis. The researchers then measured how fast and how far the DNA chasis cargoes were carried along the microtubules when different ensembles of motor molecules were attached to one DNA chasis. After determining how different number of one motor carried the cargo to one end of the microtubule and different numbers of the other motor carried the cargo to the other end of the microtubule, the researchers tried mixing both motors on one cargo chasis. Not surprisingly, in some cases no movement was observed as the two motors tugged in opposite directions. Specific cleavage of the attachments of one type of motor to the cargo released the cargo to move in the appropriate direction. In summing up their results:
Using DNA origami, we built a versatile, synthetic cargo system that allowed us to determine the motile behavior of microtubule-based motor ensembles. In ensembles of identical-polarity motors, motor number had minimal affect on directional velocity, while ensembles of opposite-polarity motors engaged in a tug-of-war resolvable by disengaging one motor species. … The system we built pro-vides a powerful platform to investigate the motile properties of any combination of identical- or opposite-polarity motors, and could also be used to investigate the role of motor regulation.
It seems likely that the system reported in this article will enable researchers to learn to regulate the movement of molecular motors carrying very diverse cargos along thick protein tracks to destinations separated by micrometers or ten of micrometers. Would such a capability contribute to the construction of useful molecular assembly lines? Could molecular motors be constructed that move cargos shorter, precisely determined distances along smaller, more precisely arranged tracks? Is this just a cute trick for learning more about biological molecular motors (after all, they do figure prominently in various serious diseases), or could this be a small step toward learning to build nanofactories?
—James Lewis, PhD