It seems that every month or two we write about another advance in structural DNA nanotechnology—the topic of the second (1995) Feynman Prize in Nanotechnology—most recently here, here, and here. DNA nanotechnology has remained important for a number of reasons. Among the specific recommendations of the 2007 Productive Nanosystems Technology Roadmap are (1) the development of modular molecular
composite nanosystems (MMCNs) in which “million-atom-scale DNA frameworks with dense arrays of distinct, addressable, [atomically precise] binding sites” provide scaffolds for organizing various nanoscale functional components (page x of Executive Summary of Productive Nanosystems: A Technology Roadmap PDF), and (2) “Prioritize modeling and design software as critical elements in the development and exploitation of [Atomically Precise Manufacturing], [Atomically Precise Productive Nanosystems], and spinoff [Atomically Precise Technologies] applications” (page ix of Executive Summary). An MIT news release written by Anne Trafton has announced major progress toward implementing both of these recommendations “Computer model enables design of complex DNA shapes“:
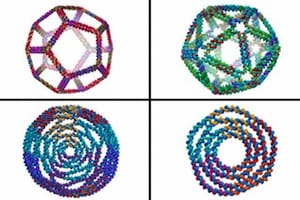
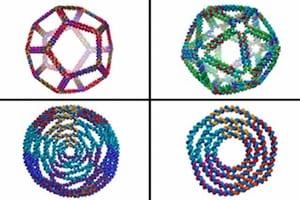
MIT biological engineers have created a new computer model that allows them to design the most complex three-dimensional DNA shapes ever produced, including rings, bowls, and geometric structures such as icosahedrons that resemble viral particles.
This design program could allow researchers to build DNA scaffolds to anchor arrays of proteins and light-sensitive molecules called chromophores that mimic the photosynthetic proteins found in plant cells, or to create new delivery vehicles for drugs or RNA therapies, says Mark Bathe, an associate professor of biological engineering.
The general idea is to spatially organize proteins, chromophores, RNAs, and nanoparticles with nanometer-scale precision using DNA. The precise nanometer-scale control that we have over 3-D architecture is what is centrally unique in this approach,” says Bathe, the senior author of a paper describing the new design approach in the Dec. 3 issue of Nature Communications [open access].
The paper’s lead authors are postdoc Keyao Pan and former MIT postdoc Do-Nyun Kim, who is now on the faculty at Seoul National University. Other authors of the paper are MIT graduate student Matthew Adendorff and Professor Hao Yan and graduate student Fei Zhang, both of Arizona State University.
DNA by design
Because DNA is so stable and can easily be programmed by changing its sequence, many scientists see it as a desirable building material for nanoscale structures. Around 2005, scientists began creating tiny two-dimensional structures from DNA using a strategy called DNA origami — the construction of shapes from a DNA scaffold” strand and smaller staple” strands that bind to the scaffold. This approach was later translated to three dimensions.
Designing these shapes is tedious and time-consuming, and synthesizing and validating them experimentally is expensive and slow, so researchers including Bathe have developed computer models to aid in the design process. In 2011, Bathe and colleagues came up with a program called CanDo that could generate 3-D DNA structures, but it was restricted to a limited class of shapes that had to be built on a rectangular or hexagonal close-packed lattice of DNA bundles.
In the new paper, Bathe and colleagues report a computer algorithm that can take sequences of DNA scaffold and staple strands and predict the 3-D structure of arbitrary programmed DNA assemblies. With this model, they can create much more complex structures than were previously possible.
The new approach relies on virtually cutting apart sequences of DNA into subcomponents called multi-way junctions, which are the fundamental building blocks of programmed DNA nanostructures. These junctions, which are similar to those that form naturally during DNA replication, consist of two parallel DNA helices in which the strands unwind and cross over,” binding to a strand of the adjacent DNA helix.
After virtually cutting DNA into these smaller sections, Bathe’s program then reassembles them computationally into larger programmed assemblies, such as rings, discs, and spherical containers, all with nanometer-scale dimensions. By programming the sequences of these DNA components, designers can also easily create arbitrarily complex architectures, including symmetric cages such as tetrahedrons, octahedrons, and dodecahedrons.
The principal innovation was in recognizing that we can virtually cut these junctions apart only to reassemble them in silico to predict their 3-D structure,” Bathe says. Predicting their 3-D structure in silico is central to diverse functional applications we’re pursuing, since ultimately it is 3-D structure that gives rise to function, not DNA sequence alone.”
The new program should enable researchers to design many more structures than those allowed by the CanDo program, says Paul Rothemund [the inventor of the scaffolded DNA origami technique and co-winner of both the Theoretical and Experimental Feynman Prizes for 2006], a senior research associate at Caltech who was not part of the research team.
Since a large fraction of the DNA nanotech community is currently using molecules whose structures could not be treated by the original CanDo, the current work is a highly welcome advance,” Rothemund says.
The researchers plan to make their algorithm publicly available within the next few months so that other DNA designers can also benefit from it. In the current version of the model, the designer has to come up with the DNA sequence, but Bathe hopes to soon create a version in which the designer can simply give the computer model a specific shape and obtain the sequence that will produce that shape. This would enable true nanometer-scale 3-D printing, where the ink” is synthetic DNA.
Scaffolds and molds
Once researchers have access to printing 3-D nanoscale DNA objects of arbitrary geometries, they can use them for many different applications by combining them with other kinds of molecules. These DNA objects are passive structural scaffolds,” Bathe says. Their function comes from other molecules attached to them.”
One type of molecule that Bathe has begun working with is light-harvesting molecules called chromophores, which are a key component of photosynthesis. In living cells, these molecules are arranged on a protein scaffold, but proteins are more difficult to engineer into nanoscale assemblies, so Bathe’s team is trying to mimic the protein scaffold structure with DNA.
Another possible application is designing scaffolds that would allow researchers to mimic bacterial toxin assemblies made from multiple protein subunits. For example, the Shiga toxin consists of five protein subunits arranged in a specific pentameric structure that enables stealthy entry into cells. If researchers could reproduce this structure, they could create a version whose toxic parts are disabled, so that the remainder can be used for delivering drugs and micro- or messenger RNAs.
This targeting subunit is very effective at getting into cells, and in a way that does not set off a lot of alarms, or result in its degradation by cellular machinery, Bathe says. With DNA we can build a scaffold for that entry vehicle part and then attach it to other things — cargo like microRNAs, mRNAs, cancer drugs, and other therapeutics.
The researchers have also used DNA nanostructures as molds to form tiny particles of gold or other metals. In a recent Science paper [see Nanodot last month], Bathe and colleagues at Harvard University’s Wyss Institute for Biologically Inspired Engineering demonstrated that DNA molds can shape gold and silver into cubes, spheres, and more complex structures, such as Y-shaped particles, with programmed optical properties that can be predicted by computer model. This approach offers a made-to-order nanoparticle design and synthesis procedure with diverse applications in nanoscale science and technology.
In this most recent paper, the authors note that atomistic modeling approaches to determining molecular shapes, such as molecular dynamics, are limited to time scales of hundreds of nanoseconds, while folding times for DNA origami typically span hours to days. On the other hand, coarse-grained finite element modeling programs, such as their earlier CanDo program, are restricted to structures based on rectangular or hexagonal lattices and cannot be applied to more general architectures, like rings and cages. To develop a computational framework to predict the 3D structure of arbitrary connections of multi-way DNA junctions, they focus here on modeling a diverse set of structures based on the ground-state geometry of immobile four-way junctions. Comparison of the modelled structures with empirical results confirmed the ability to predict 2D and 3D-curved objects that are impossible to predict using their previous CanDo system. Although important limitations remain, it is exciting that the authors intend to incorporate the new functionality into their online tool CanDo and extend its functionality to ultimately enable 3D-printing of DNA nanostructures of specified shape.
—James Lewis, PhD