A select set of videos from the 2013 Foresight Technical Conference: Illuminating Atomic Precision, held January 11-13, 2013 in Palo Alto, have been made available on vimeo. Videos have been posted of those presentations for which the speakers have consented. Other presentations contained confidential information and will not be posted.
The 3rd speaker at the Commercial Scale Devices session was Joseph Puglisi. His talk was titled “Deciphering the Molecular Choreography of Translation” biography and abstracts, video – video length 34:08.
Prof. Puglisi began with the observation that one of the triumphs of the application of chemistry and physics to biology has been the field of structural biology, his department at Stanford. Three of the last six Nobel Prizes in Chemistry have been given for determining, using primarily X-ray diffraction methods, the three-dimensional position of atoms in key molecules that drive biological function. The example that he focused upon is the ribosome—the macromolecular machine that synthesizes proteins from the genetic code. He noted, however, that one of the problems not addressed in these studies is that the pictures are static snapshots. X-ray crystallography gives you a static view of the positions of atoms. There is no motion embedded in that structure. Prof. Puglisi’s goal over the last 10-15 years has been trying to bring these structures to life—a molecular choreography to bring a time axis to structural biology to understand underlying mechanism.
The ribosome is a composite nanomaterial and a nano-object made of several RNA chains and studded with proteins. This RNA-protein complex self-assembles from RNA and protein molecules made in the cell that come together in a complex assembly pathway to form a 25 nm by 20 nm particle made of two subunits, called in bacteria the 30S and 50S subunits. It is several megadaltons in size and has binding sites for macromolecules that need to bind to the ribosome through the process of reading the genetic code.
Reviewing some basic biochemistry, Prof. Puglisi noted that messenger RNA (mRNA) has 3-letter code words that code for a given amino acid—the famous genetic code. To read the genetic code, the adapter transfer RNA (tRNA) forms base pairs with the codon within the mRNA, and has the correct amino acid specified by that codon attached to the other end of the tRNA. The ribosome positions amino acids, attached to these tRNAs through ester linkages, adjacently so that very simple chemistry occurs to form a peptide bond. An amino group attacks the carbonyl of this ester, and you’ve lengthened a peptide by one amino acid. in solution, this reaction would happen very slowly, but biology has evolved the ribosome as a catalyst to make this happen very quickly.
To achieve fidelity, the ribosome has to select the right tRNA that corresponds to the given codon and ignore the other tRNAs. Part of this fidelity is established by the base-pairing of the tRNA and the codon, but the ribosome and additional factors enhance this fidelity. The tRNAs are escorted into the ribosome by a protein factor that uses GTP as an additional source of free energy to further enhance fidelity. The tRNA that has the growing peptide chain attached to it and the incoming tRNA are positioned adjacent to one another. Steric positioning of these two groups so that the chemistry can occur accounts for a lot of the catalysis. Peptide bond formation transfers the growing peptide chain onto the tRNA that just came in.
The next step in translation is a dynamic step called translocation in which the tRNAs flux through the ribosome and the mRNA moves with respect to the ribosome. Translocation is catalyzed by another protein factor, using another GTP as a source of free energy, to precisely kick the mRNA down one codon using an unknown mechanism, to position the next codon so that the next tRNA can come in.
Prof. Puglisi noted this process has evolved to be fast and to be accurate, and that in the cell it occurs at 1 to 20 amino acids added per second. The ribosome undergoes not only conformational dynamics, but also compositional dynamics, with ligands fluxing on to the ribosome, through the ribosome, and off the ribosome. The coordination of the composition of this nanoparticle as a function of time is another key aspect of the dynamics to be explored. He noted that it is coordinated rearrangements of large domains occurring over millisecond to second timescales that drive catalysis and molecular motors in biology, so that he focuses on that timescale.
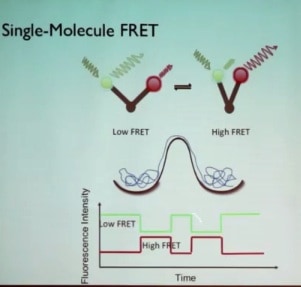
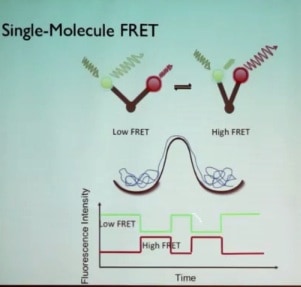
Looking for a method to track conformation as a function of time in real time, Prof. Puglisi described how that could be done using the high sensitivity of fluorophores and the high time resolution of energy transfers that occur in fluorescent spectroscopy (Förster resonance energy transfer, or FRET). If a donor fluorophore and an acceptor fluorophore are attached to a biological molecule that has two different conformers—an open form and a closed form—in which two domains shift with respect to each other, and the donor fluorophore is excited with a green laser, then non-radiative dipole-dipole coupling energy transfer to the acceptor dye occurs, and depending on the distance between the fluorophores, the acceptor dye will give off red light even though it was not excited by the laser. The transfer depends on the inverse 6th power of distance, making it very sensitive to distance. If the fluorophores are greater than 8 or 9 nm apart, no transfer occurs and all the fluorescence is green; if the fluorophores are less than 3 nm apart, all of the green light comes out as red fluorescence. The method is ideal to measure changes in distance between 4.0 and 8.0 nm, the scale of changes in the shape of the ribosome. Measuring the relative amounts of green and red light as a function of time plots the changes with time of the distance between the fluorophores, that is, the changes in conformation with time. Such changes would be averaged out looking at many molecules in solution, so meaningful results can only be obtained by looking at single molecules. This realization is the basis of Prof. Puglisi’s goal over the past 10 years: to track the dynamics of translation using single molecule methods.
Fluorescence also provides a way to track compositional dynamics, the arrival and departure of ligands. If a fluorophore is placed on a tRNA, there should be a burst of fluorescence as a ligand is bound by the ribosome, which should end when the ligand leaves. Using two different fluorophores on two different ligands gives the relative order of events by the relative appearance and disappearance of green and red light. In this way the arrival and departure of tRNAs and protein factors can be followed as components enter and leave the ribosome as translation is occurring.
To apply FRET to look at global conformational changes of the ribosome, the two different fluorophores were placed on the two different subunits to determine if the subunits rotated with respect to each other during translocation. The results (“Following the intersubunit conformation of the ribosome during translation in real time“) show that at each codon the ribosome cycles through a rotated (by about 6 degrees) and a non-rotated state upon translocation onto the next codon.”
Prof. Puglisi concluded by asking how to put together the method to study conformational dynamics and the method to study compositional dynamics. By putting one dye on the protein factor responsible for translocation, it was found that each conformational cycle correlates with the rapid arrival and departure of the protein factor. The results showed that the energy from GTP hydrolysis accelerates translocation by resetting the ribosome after peptide bond formation rotates the ribosome.
—James Lewis, PhD