While some protein scientists make impressive progress designing novel protein folds, others combine natural protein oligomers in novel ways to make unexpected extreme structures not seen in nature. A hat tip to ScienceDaily for reprinting this University of California-Los Angeles news release “UCLA biochemists build largest synthetic molecular ‘cage’ ever“:
UCLA biochemists have created the largest-ever protein that self-assembles into a molecular “cage.” The research could lead to synthetic vaccines that protect people from the flu, HIV and other diseases.
At a size hundreds of times smaller than a human cell, it also could lead to new methods of delivering pharmaceuticals inside of cells, or to the creation of new nanoscale materials.
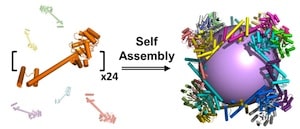
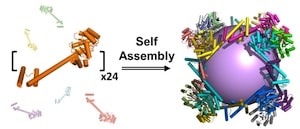
The protein assembly, which is shaped like a cube, was constructed from 24 copies of a protein designed in the laboratory of Todd Yeates, a UCLA professor of chemistry and biochemistry. It is porous — more so than any other protein assembly ever created — with large openings that would enable other large protein molecules to enter and exit.
The research was recently published online in the journal Nature Chemistry [abstract] ….
Yeates, the study’s senior author, has sought to build complex protein structures that self-assemble since he first published research on self-assembling proteins in 2001. In 2012, he and colleagues produced a self-assembling molecular cage made from 12 protein pieces combined perfectly like pieces of a puzzle. Now they have done so with 24 pieces, and they are currently attempting to design a molecular cage with 60 pieces. Building each larger protein presented new scientific challenges, but the bigger sizes could potentially carry more “cargo.”
In principle, these molecular structures should be able to carry cargo that could then be released inside of cells, said Yeates, who is a member of the UCLA–Department of Energy Institute of Genomics and Proteomics and the California NanoSystems Institute at UCLA. …
The molecular cube is probably too porous to serve as a container — for medicine, for example — inside a human body. “But the design principles for making a cage that is more closed would be the same,” Yeates said, adding that there are ways to make the cage less stable when it gets into a cell, so that it would release its cargo, such as a toxin that could kill a cancer cell.
Yeates said that his lab’s method also could lead to the production of synthetic vaccines that would mimic what a cell sees when it’s infected by a virus. The vaccines would provoke a strong response from the body’s immune system and perhaps provide better protection from diseases than traditional vaccines. …
The real work here appears to be in designing the monomer, in which two natural protein oligomers were fused using an α-helical linker, in such a way that self-assembly then joins 24 monomers to make a cube of 22.5 nm diameter, with large openings leading to a 13.0 nm diameter inner cavity. Although dwarfed by structures that can be made using DNA origami or DNA bricks or DNA cages, these large, open protein cages provide room for potentially interesting functionalization and complexity.
—James Lewis, PhD