The first journal article to call for the development of molecular manufacturing (Drexler 1981, journal publication) identified the task of designing more stable proteins as a path toward more general capabilities for molecular manipulation. Proof of principle for this goal was already apparent by 1988, and we have followed progress since then (for example, here and here). A brief comment in a recent issue of Science introduces two papers that took two different routes to use rational and computational design to make new protein structures based on alpha-helical coiled coils. In the first, a collaboration headed by David Baker, co-winner of the 2004 Foresight Feynman Prize for Theory, reported the custom design of a set of hyperstable proteins with fine-tuned geometries that can be adapted for a range of applications. From “Custom design of novel alphahelical bundles“:
Researchers at the Institute for Protein Design have developed a novel computational approach for the custom design of hyper-stable alpha-helical bundles with fine-tuned geometries. The parametric design approach and experimental characterization of the resulting helical bundles is described in detail in a recent Science publication [abstract] entitled High thermodynamic stability of parametrically designed helical bundles.
At the focus of the paper are coiled coils – a specific structural motif where two or more alpha helices twist around one another to form a supercoil. Coiled coil-based proteins play a number of important roles in biology and due to their simple and repetitive nature, have been the inspiration for a number of protein design efforts. Notably, Dr. Francis Crick proposed the concept of the coiled coil along with a set of mathematical methods for determining their structure .
To arrive at custom designed protein folds that do not exist in nature, IPD researchers and their collaborators developed a procedure for designing proteins with backbones produced by varying the parameters in the Crick coiled coil-generating equations that also utilized the Rosetta protein design methodology. Three distinct helical bundle arrangements were designed and combinatorial design calculations identified the lowest energy sequences for each supercoil arrangement. Expression and purification of these structures in the laboratory and subsequent characterization showed that these designed proteins are extremely stable and their crystal structures show very strong agreement with the design models. This new approach enables the custom design of hyperstable proteins with fine-tuned geometries for a wide range of applications including next-generation nanostructures, therapeutics, and catalysts.
The authors comment that hyperstability is easy to design for (two of nine designs tested were hyperstable) but is rarely observed among natural proteins, suggesting that “function trumped stability during natural selection.” They predict that efforts to design new protein functions will “likely move from repurposing native scaffolds to de novo design of hyperstable backbones with geometries optimal for the desired function.”
In the second paper, a team at the University of Bristol headed by Dek Woolfson demonstrated that more protein structures are possible than the thousand or so used by natural proteins. “Bristol team creates designer ‘barrel’ proteins“
This week, in the journal Science [abstract, “Computational design of
water-soluble α-helical barrels” PDF can be downloaded courtesy of corresponding author], a team of Bristol chemists and biochemists report novel, designer proteins that expand on nature’s repertoire.Proteins are long linear molecules that fold up to form well-defined 3D shapes. These 3D molecular architectures are essential for biological functions such as the elasticity of skin, the digestion of food, and the transport of oxygen in blood.
Despite the wide variety of tasks that natural proteins perform they appear to use only a limited number of structural types, perhaps just a thousand or so. These are used over and over again, being altered and embellished through evolution to generate many different functions. This begs the question: are more protein structures possible than those used and presented to us by nature?
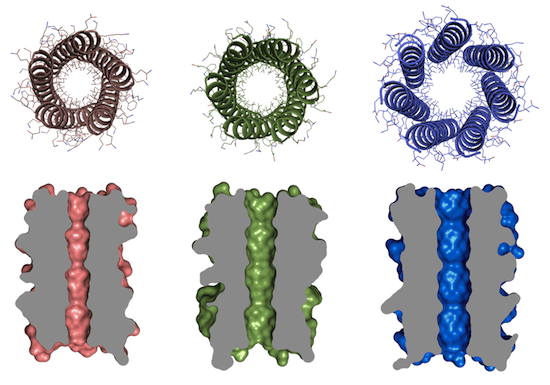
A combined team from Chemistry and Biochemistry, headed by Dek Woolfson, have addressed this by designing manmade protein molecules from scratch. Although the design principles are learnt from natural proteins, from which they developed rules for assembling the proteins, some of the designed protein shapes are completely new and have not been observed in nature yet. Specifically, they have made proteins with central cavities, or channels, running through them. The team believes that these will be useful in designing new protein functions, such as catalysts for breaking down fats, or molecules that span cell membranes to allow new communications between cells.
Dek Woolfson commented “This is a really exciting time to be exploring what can be done with biological principles and building blocks to make new and useful molecules, but completely outside the context of biology itself. It is one aspect of the emerging field of synthetic biology, in which Bristol is taking a lead both nationally and internationally.”
As explained in the second paper, α-helical barrels, a subset of coiled coils, present a promising opportunity to move beyond mimicking natural proteins. The few current examples suggest a nearly linear relationship between the number of oligomers comprising the barrel and the lumen of the barrel, but few design rules are available to design new examples. The paper describes a geometrical and computational framework for designing barrels from first principles. Although the few existing examples comprise barrels with from 5 to 12 oligomers, the authors decided to focus on rules for barrels with five to seven oligomers exemplifying ie specific type of complex coiled-coil packing. The design rules they selected identified 76 sequences, from which they selected 22 as a diverse representation for further study. Models of each were generated using various computer programs, peptides were synthesized, and cricular dichroism spectroscopy showed structures to be α-helical and thermally stable. The 20 that showed sufficient solubility were further characterized. In 8 out of 13 cases in which the oligomeric state could be determined, the result matched the computer program prediction. In four cases high-resolution x-ray crystal structures were determined, and found to closely correlate with the computer-predicted structures. The four structures produced de novo were parallel pentameric, hexameric, and heptameric structures, none of which are commonly observed in nature, and thus expanding the set of rationally designed coiled-coil assemblies. From the paper’s conclusions:
These structures have accessible and chemically defined channels, opening tantalizing prospects for the rational design of channel- and pore-containing protein assemblies with defined internal chemistries and properties …
We have commented before on the potential of synthetic biology to provide a pathway to productive nanosystems and atomically precise manufacturing/molecular manufacturing. Taken together, these two papers show that a much wider range of designed protein structures is open to exploitation for creating molecular machine systems than what can be provided solely by engineering of protein structures evolved by nature.
—James Lewis, PhD