We have frequently cited (for example, here, here, and here) the importance of modular molecular composite nanosystems (MMCNs} for the development of atomically precise productive nanosystems and eventually atomically precise manufacturing. One of the recommendations made by the 2007 Productive Nanosystems Technology Roadmap (Executive Summary, p. xi) to support the development of MMCNs is to “Develop systematic methodologies for building MMCNs in which proteins bind specific functional components to specific sites on DNA structural frameworks.” Two months ago, while researching DNA nanoswitches developed by Wesley Wong’s research group to study molecular interactions, I noted on his lab’s publications page a major contribution to achieving this objective through addressing a practical problem that could occur when making DNA-protein nanostructures “Protocol for sortase-mediated construction of DNA-protein hybrids and functional nanostructures” (open access). The abstract of the paper:
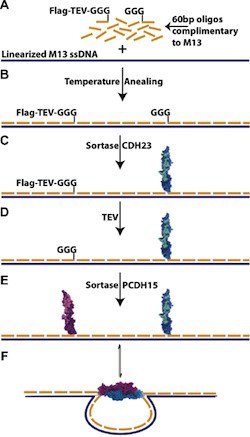
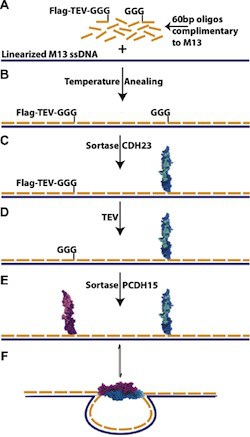
Recent methods in DNA nanotechnology are enabling the creation of intricate nanostructures through the use of programmable, bottom-up self-assembly. However, structures consisting only of DNA are limited in their ability to act on other biomolecules. Proteins, on the other hand, perform a variety of functions on biological materials, but directed control of the self-assembly process remains a challenge. While DNA–protein hybrids have the potential to provide the best-of-both-worlds, they can be difficult to create as many of the conventional techniques for linking proteins to DNA render proteins dysfunctional. We present here a sortase-based protocol for covalently coupling proteins to DNA with minimal disturbance to protein function. To accomplish this we have developed a two-step process. First, a small synthetic peptide is bioorthogonally and covalently coupled to a DNA oligo using click chemistry. Next, the DNA–peptide chimera is covalently linked to a protein of interest under protein-compatible conditions using the enzyme sortase. Our protocol allows for the simple coupling and purification of a functional DNA–protein hybrid. We use this technique to form oligos bearing cadherin-23 and protocadherin-15 protein fragments. Upon incorporation into a linear M13 scaffold, these protein–DNA hybrids serve as the gate to a binary nanoswitch. The outlined protocol is reliable and modular, facilitating the construction of libraries of oligos and proteins that can be combined to form functional DNA–protein nanostructures. These structures will enable a new class of functional nanostructures, which could be used for therapeutic and industrial processes.
The introduction to the paper points out that many chemistries conventionally used to link proteins and DNA react with functional groups that are ubiquitous in biology so that the DNA oligo could react to amy of several sites on the protein, increasing the risk of interfering with protein function. This risk is avoided by coupling the DNA oligo to a small, synthetic peptide, so there is no need to optimize coupling protocols for each protein. The protein of interest is then coupled to this DNA-peptide chimera using sortase, a microbial enzyme that recognizes a short amino acid sequence LPX1TGX2 near the C-terminus of one protein and couples it to a GGG sequence at the N-terminus of another protein to produce a fusion of the two proteins. The variant of sortase used was evolved to optimize coupling under conditions favorable for protein stability. Thus all of the chemistry that might be damaging to proteins is front-loaded so that is performed on a small peptide designed to be compatible with those conditions. The peptide can also be designed to facilitate easy purification of the DNA oligo-synthetic peptide-fusion protein. The end result of this process is that it is straight-forward to systematically build a library of proteins linked to DNA oligos, which can then be used to bind any of those proteins to any desired site on a DNA origami structure.
The authors present detailed protocols to (1) form a DNA-oligo bearing a peptide compatible with the sortase enzyme, (2) the sortase-catalyzed coupling of a protein to the DNA-peptide chimera, and (3) the integration of the DNA-protein hybrids into self-assembling nanostructures. Two different self-assembly protocols were presented, depending on whether or not the protein would remain stable at 40° C.
In concluding, the authors note:
When combined with a library of sortase compatible oligos and peptides, this flexible and modular approach could enable the creation of a wide range of functional nanostructures on demand. Furthermore, our approach expands the range of functional DNA–protein chimeras that can be constructed, enabling the incorporation of previously inaccessible protein machinery to generate nanostructures with previously unobtainable functionalities.
It will be of interest to see if “previously unobtainable functionalities” can be identified that will accelerate the development of productive nanosytems.
—James Lewis, PhD