More evidence that computational protein design can create not only novel proteins but also novel functions that do not exist in nature comes from the creation of an entire novel metabolic pathway. A large collaboration involving scientists from the University of California, Davis, two research groups at the University of Washington (including the lab of David Baker, who shared the 2004 Foresight Institute Feynman Prize for theoretical work), the Fred Hutchinson Cancer Research Center, and several other institutions in California and Israel published a paper last year in PNAS “Computational protein design enables a novel one-carbon assimilation pathway” that describes a novel computationally designed enzyme they designate “formolase” that catalyzes the carboligation of three one-carbon formaldehyde molecules into one three-carbon dihydroxy acetone molecule. This complex project comprised many steps to create three novel enzyme functions, not previously known, in the process creating a microbial metabolic pathway that could be further optimized for enhanced production of desired products. This research demonstrates the feasibility of organizing multiple engineered enzymes into a sequence that does not exist in nature. In this particular case the goal is to address current challenges in energy storage and carbon sequestration by converting one carbon compounds, such as CO2, into multicarbon fuels and other high-value chemicals. One could also envision such systems as components along the path to productive nanosystems, leading eventually to general purpose, high throughput atomically precise manufacturing.
The authors note that the lack of one-carbon anabolic pathways in microbes suitable to address current needs in energy storage and carbon sequestration could arise from unfavorable chemical driving force at one or more pathway steps, the inherent complexity and inefficiency of the steps, or the environmental sensitivity of the steps (the ability to function efficiently under both aerobic and anaerobic conditions). Despite the lack of such a pathway in nature, they further note, the established electrochemical reduction of CO2 to formate provides an attractive starting point for a one-carbon fixation pathway. They describe in this paper the computational design of an enzyme that catalyzes the carboligation of three one-carbon formaldehyde molecules into one three-carbon molecule. The new enzyme enables the construction of the ‘formolase’ pathway, which converts formate into the centrally important metabolite dihydroxyacetone phosphate.
To find a place to start, the authors note that although there is no biochemical precedent for coupling formate molecules into a multicarbon molecule, there is a well-known organic synthesis (the formose reaction) for coupling formaldehyde into dihydroxyacetone. Although no enzyme is known to catalyze this formose reaction, the observation that thiazolium salts were known to catalyze the formose reaction provided an initial clue, pointing toward thiamine pyrophosphate-dependent enzymes. The enzyme benzaldehyde lyase (BAL) was identified as a promising starting point because it catalyzes a related coupling of two benzaldehyde molecules to form one benzoin molecule.
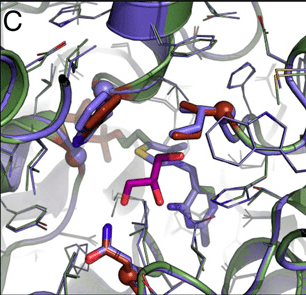
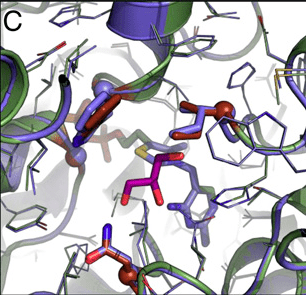
The authors synthesized and expressed the gene for benzaldehyde lyase, purified the protein and assayed it for formose activity. A small amount of formose activity was found—about 36,000-fold lower than for its natural reaction with benzaldehyde. They then used computational protein design to redesign the benzaldehyde binding pocket of the enzyme to increase formose activity. Four cycles of computational design and experimental evaluation totaling 121 designs led to a variant they designated Des1 with 26-fold higher activity and four amino acid substitutions. The crystal structure of Des1 showed that the computational modeling accurately predicted the structure of the active site of the protein. One additional round of computationally guided site-directed mutagenesis and two mutations found through error-prone PCR further increased Des1 activity 4-fold. Crystalizing and solving the structure of this new 7-site mutant, termed formolase, showed the overall conformation of the protein backbone and active site was similar to the Des1 intermediate step. Formolase showed a 100-fold increase in formose activity compared to the original BAL enzyme, and a 100,000-fold decrease in benzoin activity, resulting in a total specificity switch of greater than 10 million-fold.
An active formolase provides the opportunity to design a pathway to convert formate to dihydroxyacetone phosphate. However, the first step, the direct reduction of formate to formaldehyde using the metabolite NADH is an extremely unfavorable reaction. To accomplish this reaction, the authors sought to activate formate with Coenzyme A to form formyl-CoA, thus reducing the thermodynamic barrier to reducing formate to formaldehyde by NADH. No enzyme is known to catalyze this reaction, so an enzyme that catalyzes the similar reaction with acetate was cloned, expressed, and purified. Assaying for activity of this enzyme with formate as substrate revealed a clear signal.
The second step of the pathway is the reduction of formyl-CoA to formaldehyde. No enzyme is known to catalyze this step either, but enzymes are know that catalyze the reversible reduction of acetyl-CoA to acetaldehyde. The first of these enzymes tested had little activity to produce formyl-CoA, but genes for five homologous enzymes were identified, synthesize, expressed, purified and tested, and one was 100-fold more active than the first one tested. Combining the enzymes identified for the first and second steps reduced formate to formaldehyde.
Combining the above two enzymes with the de novo designed formolase and a couple of known enzymes produced a complete pathway from formate to dihydroxyacetone phosphate. Two rate-limiting steps were identified to be targeted for future enzyme engineering efforts
The formolase enzyme designed here and the two novel activities of existing enzymes identified here provide a potential route for biocatalytic conversion of one-carbon molecules into central metabolites. The authors note that this formolase pathway compares favorably with the nine known naturally occurring pathways to utilize formate or carbon dioxide. THe five steps required are fewer than all the natural pathways, it functions under fully aerobic conditions, and it has a higher chemical driving force than any of the natural pathways. THe authors discuss ways in which the pathway could be further improved, and how it could be extended to use CO2 as a sole carbon and energy source through the electrochemical reduction of CO2 to formate.
This impressive body of work provides a proof-of-principle demonstration of the ability of protein design to extend the reach of synthetic biology to address current challenges using proteins newly designed specifically for those challenges, rather than being limited to recombining natural proteins optimized by evolution for other challenges.
—James Lewis, PhD