Scaffolded DNA origami provided the starting point for the modular molecular composite nanosystems approach to atomically precise productive nanosystems (see Productive Nanosystems: A Technology Roadmap PDf page x, page 51). Over the past seven years we have pointed here to numerous advances in structural DNA nanotechnology and the related underlying technologies as advances toward atomically precise manufacturing, but one limit has been the maximum size available of the conventional single stranded scaffold available from the bacteriophage M13, which limits the scale of the uniquely addressable DNA structures that can be built. A second limit has been the cost of synthesis of the DNA staple strands. Now both of those limits have been greatly extended. A hat tip to ScienceDaily for reprinting this North Carolina State University news release “Researchers Create World’s Largest DNA Origami“:
Researchers from North Carolina State University, Duke University and the University of Copenhagen have created the world’s largest DNA origami, which are nanoscale constructions with applications ranging from biomedical research to nanoelectronics.
“These origami can be customized for use in everything from studying cell behavior to creating templates for the nanofabrication of electronic components,” says Dr. Thom LaBean, an associate professor of materials science and engineering at NC State and senior author of a paper describing the work.
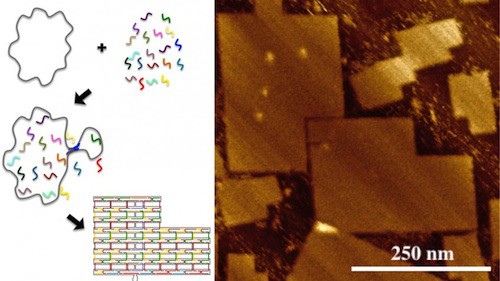
DNA origami are self-assembling biochemical structures that are made up of two types of DNA. To make DNA origami, researchers begin with a biologically derived strand of DNA called the scaffold strand. The researchers then design customized synthetic strands of DNA, called staple strands. Each staple strand is made up of a specific sequence of bases (adenine, cytosine, thaline [sic] [they meant “thymine”] and guanine — the building blocks of DNA), which is designed to pair with specific subsequences on the scaffold strand.
The staple strands are introduced into a solution containing the scaffold strand, and the solution is then heated and cooled. During this process, each staple strand attaches to specific sections of the scaffold strand, pulling those sections together and folding the scaffold strand into a specific shape.
The standard for DNA origami has long been limited to a scaffold strand that is made up of 7,249 bases, creating structures that measure roughly 70 nanometers (nm) by 90 nm, though the shapes may vary.
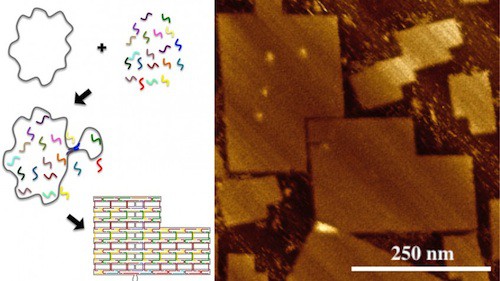
However, the research team led by LaBean has now created DNA origami consisting of 51,466 bases, measuring approximately 200 nm by 300 nm.
“We had to do two things to make this viable,” says Dr. Alexandria Marchi, lead author of the paper and a postdoctoral researcher at Duke. “First we had to develop a custom scaffold strand that contained 51 kilobases. We did that with the help of molecular biologist Stanley Brown at the University of Copenhagen.
“Second, in order to make this economically feasible, we had to find a cost-effective way of synthesizing staple strands — because we went from needing 220 staple strands to needing more than 1,600,” Marchi says.
The researchers did this by using what is essentially a converted inkjet printer to synthesize DNA directly onto a plastic chip.
“The technique we used not only creates large DNA origami, but has a fairly uniform output,” LaBean says. “More than 90 percent of the origami are self-assembling properly.”
The paper, “Toward Larger DNA Origami,” is published online in Nano Letters [abstract]. …
These workers used very clever genetic engineering to produce a hybrid of a double-stranded DNA bacteriophage and the single-stranded DNA bacteriophage traditionally used to produce a scaffold strand, thus producing a scaffold about seven times longer than the conventional one, enabling the production of an origami with a 9.5-fold greater area. Even more useful than this accomplishment was the use of a converted inkjet printer to lower the cost of staple strands from $0.35 per base to $0.001 per base. Without this large drop in price, the cost of synthesizing staples for a larger scaffold would have been prohibitive for most budgets. In fact, the cost of the staple strands was a major deterrent for wider use of the technology even with the smaller, conventional scaffold. This advance provides an example of why it can be difficult to predict the rate of technological advance toward an ambitious, difficult goal. Incremental advances using related technologies can suddenly make an anticipated path more practical than anticipated. The development of larger scaffolds could perhaps have been easily predicted by someone familiar with genetic engineering. The 350-fold drop in the cost of synthesizing DNA staples was perhaps more surprising.
—James Lewis, PhD