High-throughput atomically precise manufacturing (APM) has been described as a manufacturing technology that could be developed over the next few decades that could radically change civilization (Radical Abundance). APM has been alternatively referred to as “molecular nanotechnology”, “molecular manufacturing” and “productive nanosystems”.
For a very limited class of objects, chemists have been doing APM for two centuries by synthesizing small to mid-size molecules to atomic precision (plus large molecules with repeating subunits, like polymers and dendrimers). In the case of molecules without repeating subunits, the limits of synthetic chemistry are reached when molecules become large enough that chemists cannot devise methods to add the next group of atoms at one specific site on the growing molecule instead of at other chemically similar sites. In proposals to develop APM, this problem will be solved by guided molecular trajectories and positionally-controlled mechanosynthesis: reactive molecular fragments will be guided by molecular machine systems to a specific position and geometric orientation with respect to the growing atomically precise work piece so that the application of mechanical force will cause the desired bond to form.
APM has also been likened to an atomic scale version of 3D-printing (also called additive manufacturing) in which a small volume element (but a couple orders of magnitude larger than molecular scale) can be added at any arbitrary position to a growing object (see, for example, Radical Abundance pp 76-77). Thus APM and 3D-printing are both manufacturing systems in which arbitrarily complex 3D objects can be manufactured from the bottom-up through the positionally controlled placement of very small elements.
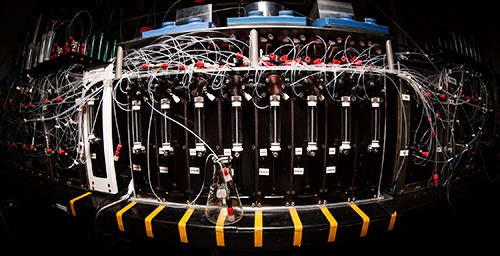
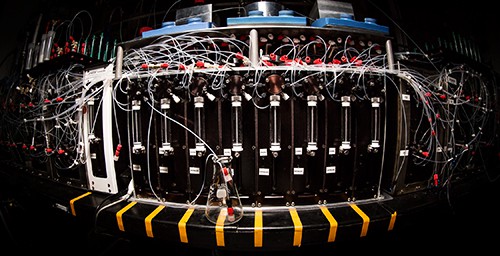
In recent weeks, the blogosphere has greeted a radical advance in synthetic organic chemistry with titles like “3-D Printer for Small Molecules Opens Access to Customized Chemistry” (Phys Org), “Scientists Unveil 3D Printer for Small Molecules” (STGIST), and “This Chemistry 3D Printer Can Synthesize Molecules from Scratch”. To what extent does this huge advance in synthetic chemistry technology amount to bringing the positional control of 3D-printing to the molecular scale? How close does it get us to APM? From the University of Illinois news bureau “Molecule-making machine simplifies complex chemistry”:
A new molecule-making machine could do for chemistry what 3-D printing did for engineering: Make it fast, flexible and accessible to anyone.
Chemists at the University of Illinois, led by chemistry professor and medical doctor Martin D. Burke, built the machine to assemble complex small molecules at the click of a mouse, like a 3-D printer at the molecular level. The automated process has the potential to greatly speed up and enable new drug development and other technologies that rely on small molecules.
“We wanted to take a very complex process, chemical synthesis, and make it simple,” said Burke, a Howard Hughes Medical Institute Early Career Scientist. “Simplicity enables automation, which, in turn, can broadly enable discovery and bring the substantial power of making molecules to nonspecialists.”
The researchers described the technology in a paper featured on the cover of the March 13 issue of Science. [abstract, full text PDF courtesy of Burke Lab]
“Small molecules” are a specific class of complex, compact chemical structures found throughout nature. They are very important in medicine – most medications available now are small molecules – as well as in biology as probes to uncover the inner workings of cells and tissues. Small molecules also are key elements in technologies like solar cells and LEDs.
However, small molecules are notoriously difficult to make in a lab. Traditionally, a highly trained chemist spends years trying to figure out how to make each one before its function can even be explored, a slowdown that hinders development of small-molecule-based medications and technologies.
“Up to now, the bottleneck has been synthesis,” Burke said. “There are many areas where progress is being slowed, and many molecules that pharmaceutical companies aren’t even working on, because the barrier to synthesis is so high.”
The main question that Burke’s group seeks to answer: How do you take something very complex and make it as simple as possible?
The group’s strategy has been to break down the complex molecules into smaller building blocks that can be easily assembled. The chemical building blocks all have the same connector piece and can be stitched together with one simple reaction, the way that a child’s interconnecting plastic blocks can have different shapes but all snap together. Many of the building blocks Burke’s lab has developed are available commercially.
See a video of Burke explaining the process.
To automate the building-block assembly, Burke’s group devised a simple catch-and-release method that adds one building block at a time, rinsing the excess away before adding the next one. They demonstrated that their machine could build 14 different classes of small molecules, including ones with difficult-to-manufacture ring structures, all using the same automated building-block assembly.
“Dr. Burke’s research has yielded a significant advance that helps make complex small molecule synthesis more efficient, flexible and accessible,” said Miles Fabian of the National Institutes of Health’s National Institute of General Medical Sciences, which partially funded the research. “It is exciting to think about the impact that continued advances in these directions will have on synthetic chemistry and life science research.”
The automated synthesis technology has been licensed to REVOLUTION Medicines, Inc., a company that Burke co-founded that focuses on creating new medicines based on small molecules found in nature. The company initially is focusing on anti-fungal medications, an area where Burke’s research has already made strides.
“It is expected that the technology will similarly create new opportunities in other therapeutic areas as well, as the industrialization of the technology will help refine and broaden its scope and scalability,” Burke said.
“Perhaps most exciting, this work has opened up an actionable roadmap to a general and automated way to make most small molecules. If that goal can be realized, it will help shift the bottleneck from synthesis to function and bring the power of making small molecules to nonspecialists.”
This paper notes that complex natural products are biosynthesized via the iterative assembly of a small set of building blocks. The researchers used an approach analogous to iterative peptide coupling that sequentially assembles bifunctional N-methyliminodiacetic acid (MIDA) boronates. The coupling is basically a Suzuki coupling reaction (in which an organic compound carrying a boronate and another organic compound carrying a halogen are joined by a Pd catalyst, creating a new C-C single bond joining the two organic fragments and spitting out the boronate and the halogen). The MIDA group covers the boron on one partner to make it unavailable, analogous to the protective group strategy used in solid phase peptide synthesis to insure that each cycle only forms one coupling.
In this approach all of the functional groups, oxidation states, and stereochemical elements required to get a diverse array of products are preinstalled into building blocks. The major challenge the authors faced was to include cyclic and polycyclic natural products rich in sp3-hybridized carbon atoms. Because many natural products of complex topology are biosynthesized by cyclization of modular linear precursors, which are in turn derived from iterative building block assembly, the authors hypothesized that an analogous linear to cyclized strategy could be used to access all fourteen types of structures presented here.
To automate this process, it was next necessary to devise a common purification scheme that could be used for all syntheses. In the case of peptide, oligonucleotide, and certain other polymer syntheses, the need for purification after each step is addressed by solid phase synthesis. However, a different solution is required for small molecules because they lack a common functional group that can be employed for attachment to the solid phase. Since each iteration of their building block assembly generates a MIDA boronate as the key intermediate, they looked at whether this motif could serve as a common handle for purification. The discovery that a very wide variety of MIDA boronate intermediates would bind tightly to silica with a methanol/diethyl ether solvent, but elute rapidly with tetrahydrofuran provided a catch-and-release purification protocol. The authors then designed and built a fully automatic synthesizer that sequentially executes deprotection, coupling, and purification steps for each cycle of the synthesis. They were also able to add carbon-heteroatom bond formation to the same synthetic platform.
A feature article in the same issue of Science provides a perspective on this research “The Synthesis Machine: An automatic device that makes small organic molecules could revolutionize drug discovery” (full text courtesy of Burke Lab).) Interesting numbers on the capability of this approach:
- About 260,000 small molecule natural products are known
- 70-75% of those could be made with 5000 MIDA boronate building blocks
- About 200 MIDA building blocks are now commercially available
The main limitation right now is that only 200 building blocks are available that carry both the halogen and the boronate groups, so only these can be used anywhere other than the first or last building block in the chain.
If several thousand new building blocks become commercially available, it may not be too much of an exaggeration to talk about 3D printing of small molecules. The biggest difference is that you cannot use simple positional control to arbitrarily place the next element of the growing molecule, as you can with the next element of a 3D-printed structure. Rather, you have to design whatever elements of structural information are necessary to get the right structure into your choice of one or more building blocks so that after the linear precursor goes through the final cyclization cascade, the pieces will all be where they should be. In some ways this is reminiscent of protein design, in which a sequence of amino acids has to be determined that will fold into the desired 3D structure. But in this case the choice of subunits is vastly greater. Will this make the design process easier or more difficult as the products become larger?
It will no doubt take several years to see just how big, how diverse, and how complex the products of this molecule synthesizer can be. It seems reasonable to expect a huge effect on drug discovery and other practical areas. Will it contribute to the development of APM? For example, could it synthesize small diamondoid parts? Could it produce useful catalysts or other functional components to attach to a large structural DNA framework? Or perhaps components for smaller, stiffer structural frameworks? Let’s hope that this machine does indeed become widely available as Prof. Burke hopes so that we can see just how useful it can become.
—James Lewis, PhD