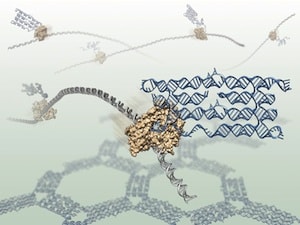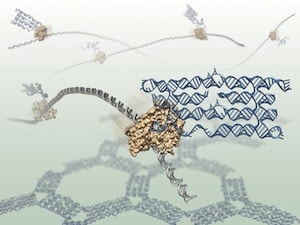
We have drawn attention here to RNA nanotechnology because RNA can function as a genetic material, as can DNA, so it has a molecular recognition code similar (but not identical) to that of DNA, but it can also assume a wider range of secondary structures than can DNA, and thus can also function catalytically, as do proteins. Since the replication and transmission of genetic information and the catalysis of chemical reactions are the two most basic functions of the molecular machinery of life, it is of considerable interest to explore what role RNA nanotechnology could play in developing the artificial molecular machine systems that will lead both to near term advances in various areas of nanotechnology and to the ultimate development of high-throughput atomically precise manufacturing (APM). Structural DNA nanotechnology predates RNA nanotechnology by a decade or more, and one of key developments in building more complex nanostructures and devices from DNA has been DNA origami. So, what might we expect from last summer’s introduction of RNA origami? A hat tip to ScienceDaily for reprinting this news release from Aarhus University, and for adding an extremely useful “RNA fact sheet” at the bottom of the article that summarizes the key roles of RNA and the differences in structure and function between RNA and DNA. “Scientists Fold RNA Origami From a Single Strand“:
RNA origami is a new method for organizing molecules on the nanoscale. Using just a single strand of RNA, many complicated shapes can be fabricated by this technique. Unlike existing methods for folding DNA molecules, RNA origamis are produced by enzymes and they simultaneously fold into pre-designed shapes. These features may allow designer RNA structures to be grown within living cells and used to organize cellular enzymes into biochemical factories. The method, which was developed by researchers from Aarhus University (Denmark) and California Institute of Technology (Pasadena, USA), is reported in the latest issue of Science [abstract].
Origami, the Japanese art of paper folding, derives its elegance and beauty from the manipulation of a single piece of paper to make a complex shape. The RNA origami method described in the new study likewise involves the folding of a single strand of RNA, but instead of the experimenters doing the folding, the molecules fold up on their own.
“What is unique about the method is that the folding recipe is encoded into the molecule itself, through its sequence,” explains Cody Geary, a postdoctoral scholar in the field of RNA structure and design at Aarhus University.
“The sequence of the RNAs defines both the final shape and also the series of movements that rearrange the structures as they fold.”
“The challenge of designing RNAs that fold up on their own is particularly difficult, since the molecules can easily get tangled during the folding process. So to design them, you really have to imagine the way that the molecules must twist and bend to obtain their final shape,” Geary says.
Nano size honeycombs
The researchers used 3D models and computer software to design each RNA origami, which was then encoded as a synthetic DNA gene. Once the DNA gene was produced, simply adding the enzyme RNA-polymerase resulted in the automatic formation of RNA origami.
To observe the RNA molecules, the researchers used an atomic force microscope, a type of scanning microscope that softly touches molecules instead of looking at them directly. The microscope is able to zoom in a thousand times smaller than is possible with a conventional light microscope. The researchers demonstrated their method by folding RNA structures that form honeycomb shapes, but many other shapes should be realizable.
“We designed the RNA molecules to fold into honeycomb patterns because they are easy to recognize in the microscope. In one experiment we caught the polymerases in the process of making the RNAs that assemble into honeycombs, and they really look like honey bees in action,” Geary continues. /p>
Better and cheaper
A method for making origami shapes out of DNA has been around for almost a decade, and has since created many applications for molecular scaffolds. However, RNA has some important advantages over its chemical cousin DNA that make it an attractive alternative:
Paul Rothemund, a research professor at the California Institute of Technology and the inventor of the DNA origami method, is also an author on the new RNA origami work.
“The parts for a DNA origami cannot easily be written into the genome of an organism. RNA origami, on the other hand, can be represented as a DNA gene, which in cells is transcribed into RNA by a protein machine called RNA polymerase,” explains Rothemund.
Rothemund further adds, “The payoff is that unlike DNA origami, which are expensive and have to be made outside of cells, RNA origami should be able to be grown cheaply in large quantities, simply by growing bacteria with genes for them. Genes and bacteria cost essentially nothing to share, and so RNA origami will be easily exchanged between scientists.”
Self-assembling scaffolds
The research was performed at laboratories at Aarhus University in Denmark, and the California Institute of Technology in Pasadena. Ebbe Andersen, an Assistant Professor at Aarhus University, who works on developing molecular biosensors, led the development of the project.
“All of the molecules and structures that form inside of living cells are the products of self-assembly, but we still know very little about how self-assembly actually works. By designing and testing self-assembling RNA shapes, we have begun to shed some light on fundamental principles of self-assembly,” says Andersen.
“The primary application for these molecular shapes is to build scaffolds for arranging other microscopic components, such as proteins, into groups that allow them to work together. For example, using the scaffolds as a foundation to build a microscopic chemical factory in which products are passed from one protein enzyme to the next,” Andersen explains. …
A Caltech news release included some additional details:
… So far, the researchers have demonstrated their method by designing RNA molecules that fold into rectangles and then further assemble themselves into larger honeycomb patterns. This approach was taken to make the shapes recognizable using an atomic force microscope, but many other shapes should be realizable. …
The particular RNA sequences that were folded in the experiment were designed using software called NUPACK, created in the laboratory of Caltech professor Niles Pierce. Both the Rothemund and Pierce labs are funded by a National Science Foundation Molecular Programming Project (MPP) Expeditions in Computing grant.
“Our latest research is an excellent example of how tools developed by one part of the MPP are being used by another,” says Rothemund. …
“RNA origami is still in its infancy,” says Rothemund. “Nevertheless, I believe that RNA origami, because of their potential to be manufactured by cells, and because of the extra functionality possible with RNA, will have at least as big an impact as DNA origami.” …
An additional Insights piece in the same issue of Science (“Self-assembled RNA nanostructures” by Neocles B. Leontis and Eric Westhof) provides additional information on the variety of non-Watson-Crick base pairs in RNA and how these provide additional structural modules that serve as building blocks of tertiary architecture of folded RNA transcripts. This parallels the process in which protein secondary structure motifs form the components of the tertiary structure of folded protein molecules. As with protein structures, the folder structures of many large RNA molecular machines are known in atomic detail. They note that:
… One of the holy grails of RNA nanotechnology has been to rationally design objects comparable in size and complexity to natural RNA machines like the ribosome and spliceosome, as well as current highly sophisticated DNA nanostructures. …
The results presented in this debut of RNA origami demonstrate the formation of RNA tertiary motifs to construct hexagonal tiles that assemble into honeycomb lattices. Different variants could self-assemble cotranscriptionally or on a mica surface, or both. The largest tiles demonstrated were 660 nucleotides in length. So far, RNA origami scaffolds that match the size of the largest DNA origami structures (51,466 bases, approximately 200 nm by 300 nm) can only be made from the self-assembly of dozens of identical tiles. However, the expected basic advantages of cotranscriptional folding, which could lead to in vivo fabrication of RNA origami structures for use in the cell, and for inexpensive production of large quantities, have been demonstrated. Also, the greater variety and versatility of RNA motifs for 3D structures, finer detail, and potential for functional nanomachines remain to be systematically exploited. However, the greater complexity of RNA structures and functions compared to DNA makes design more challenging, as is the case for protein design. Much remains to be done, but the number of options for the productive nanosystems approach to APM seems to be growing.
—James Lewis, PhD