We have frequently cited examples of the artificial molecular machines that can be built from DNA. An open question is whether these prototype molecular machines can be improved toward practical applications. For example, can simple machines for manipulating molecules be improved to the point of implementing atomically precise manufacturing? A recent publication provides an example of rational improvement of a simple DNA machine reported three years ago. Three years ago a news release from the Biodesign Institute of Arizona State University reported “Tiny tweezers allow precision control of enzymes“:
Tweezers are a handy instrument when it comes to removing a splinter or plucking an eyebrow.
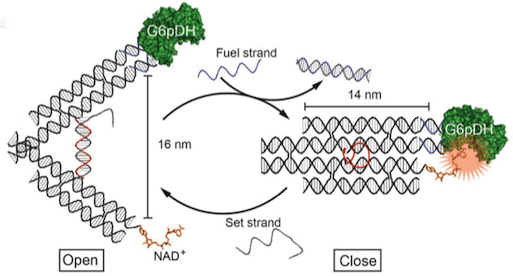
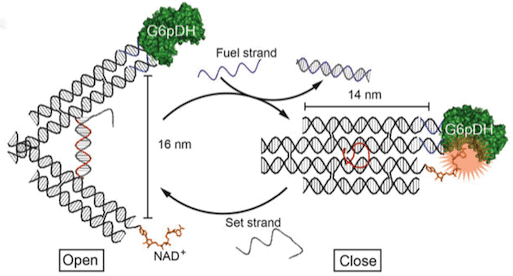
In new [2013] research, Hao Yan and his colleagues at Arizona State University’s Biodesign Institute describe a pair of tweezers shrunk down to an astonishingly tiny scale. When the jaws of these tools are in the open position, the distance between the two arms is about 16 nanometers—over 30,000 times smaller than a single grain of sand.
The group demonstrated that the nanotweezers, fabricated by means of the base-pairing properties of DNA, could be used to keep biological molecules spatially separated or to bring them together as chemical reactants, depending on the open or closed state of the tweezers.
In a series of experiments, regulatory enzymes—central components in a host of living processes—are tightly controlled with the tweezers, which can switch reactions on or off depending on their open or closed condition.
“The work has important implications for regulating enzymatic function and may help usher in a new generation of nanoscale diagnostic devices as well as aid in the synthesis of valuable chemicals and smart materials”, said Yan.
Results of the new research appear in the current issue of the journal Nature Communications “A DNA tweezer-actuated enzyme nanoreactor“. …
Enzymes are large molecules responsible for thousands of chemical interactions essential to life. A primary role for enzymes is to accelerate or catalyze myriad chemical reactions involved in processes ranging from digestion to DNA synthesis. To do this, enzymes lower the activation energy—the minimum energy needed for chemical reactions to occur—thereby speeding up the rate of such reactions. Enzymes are critical factors for health and disease, helping cells maintain their delicate homeostasis. When mutations lead to over- or under-production in certain key enzymes, severe genetic diseases—some of them, lethal—can result.
Because of the central importance of enzymes for biological systems, researchers want to gain a better understanding of how normal enzymatic reactions occur and how they may go awry. Such knowledge may encourage the development of techniques to mimic cellular processes involved in enzyme regulation.
In the current study, the authors create a nanoscale tool designed to manipulate enzymatic reactions with fine-grained control. The group dubs their device a tweezer-actuated enzyme nanoreactor.
The clever design separates an enzyme and a cofactor essential for successful reactions on separate arms of the tweezer-like instrument (See Figure 1). Enzyme function is inhibited when the tweezers are in their open position and the two molecules are held apart. Enzyme activation takes place when the tweezer prongs close, bringing enzyme and cofactor in contact. (The closing of the tweezers occurs when a specific DNA sequence is added, altering the thermodynamics of the system and causing a conformational change in the structure.)
The current study explores reactions in regulatory enzymes—multitasking entities that are important for modulating biochemical pathways. Regulatory enzymes, which can catalyze reactions over and over again, accomplish their feats by binding with biomolecular cofactors. (Hormone production and regulation are just one example of regulatory enzyme activity.)
In a series of experiments, the group was able to externally control the inhibition and activation of the enzyme through successive cycles. The authors stress that the nanoreactor tweezers could be used to regulate other types of enzymes and their control could be further refined by means of feedback and feed forward loops.
Engineering nanostructures from the bottom up, using DNA as a construction material, affords researchers exacting control over the resulting geometry. Previously, Yan has created nanostructures in two and three-dimensions, ranging from flat shapes to bowls, baskets, cages, Möbius strips and a spider-like autonomous walker.
In the tweezer design, a pair of 14 nm arms is connected at their ends by means of a 25 nucleotide single strand of DNA. This strand controls the opening and pinching of the tweezers, much the way a spring acts in a pair of gardening shears.
Two types of complementary sequence strand interact with this component, either forming a rigid DNA double helix, which supports the tweezers in their open position (set strands) or disabling the structural support and closing the tweezers, (fuel strands).
Two techniques were used to measure and analyze the resulting structures with nanoscale precision: Fluorescence Resonance Energy Transfer (FRET) and Atomic Force Microscopy (AFM). Experiments demonstrated a high yield for enzyme-bound tweezers, and successful switching between open and closed states was observed. The use of FRET allowed the process to be monitored in real time.
Lengthening the cofactor linker dangling from one of the tweezer’s arms enhanced successful opening and closing of the enzyme tweezers. Analysis revealed a 5-fold increase in enzymatic activity in the closed state compared with the open state. The study also demonstrated durability in the tweezers, which were able to cycle between the open and closed positions 9 times without losing structural integrity. The process was only limited by the accumulation of set strands and fuel strands.
Future work will explore similar responsive enzyme nanodevices capable of selective chemical amplification, with potentially broad impacts for medical diagnostics. Nanoreactors may also be applied as precision biocatalysts for the production of useful chemicals and smart materials.
The recent (2016) paper, from the Arizona State group working with groups from the University of Michigan and the Massachusetts Institute of Technology, endeavors to improve the relatively modest six-fold enhancement between open and closed tweezers positions seen in the first paper: “Rational design of DNA-actuated enzyme nanoreactors guided by single molecule analysis” (abstract, PDF made available by one of authors). To understand the structure-function relationships in the DNA tweezers, the authors combined molecular dynamics simulations with quantitative single molecule imaging using fluorescence resonance energy transfer (FRET) and atomic force microscopy (AFM). These experiments revealed two important deficiencies in the tweezers design: incomplete closure upon actuation, and conformational heterogeneity. Rational redesign of the DNA crossover motifs located in the hinge and arms regions led to more complete and more uniform closures.
FRET experiments with the initial tweezers designed showed one conformation in the open state, as expected, but two distinct conformations in the closed state, corresponding to distances between the two arms of > 10 nm and ~6.3 nm, respectively. Molecular dynamics simulations showed that incomplete closure was due to a flexible spacer of three thymidine residues. Eliminating the three-thymidine spacer, however, led to even less closure through the introduction of twisting between the arms of the tweezers. Switching to a one-thymidine spacer and re-designing the actuating hairpin by slightly extending the stem to increase stability improved performance, but did not eliminate heterogeneity.
Using the original enzyme plus cofactor activity assay to compare the original design with two of the above improved designs, one was found to be twice as active and the other three times as active. Single molecule observations suggest the presence of:
… two different mechanisms for improving nanoreactor performance: (i) a greater number of nanoreactors reach the critical [interarm] distance needed for activating their coupled enzyme with its cofactor due to improved closure upon optimized design of the actuator hairpin; and (ii) more frequent substrate turnover is achieved when stabilizing interarm tail–tail interactions are implemented that lead to straighter tweezer arms resulting from improved [Holliday junction] sequence design.
This research identified specific features of the original tweezers design that resulted in suboptimal performance and implemented a rational design strategy to improve each of two structural elements. The authors are confident that because the features identified here are general to many DNA nanostructures, they have established guidelines to rationally improving performance of a variety of nanodevices. It will be interesting to see how widely applicable these guidelines are, how far they can take performance improvement of various devices, and what new devices or systems such improvements make possible.
—James Lewis, PhD
Discuss these news stories on Foresight’s Facebook page or on our Facebook group.