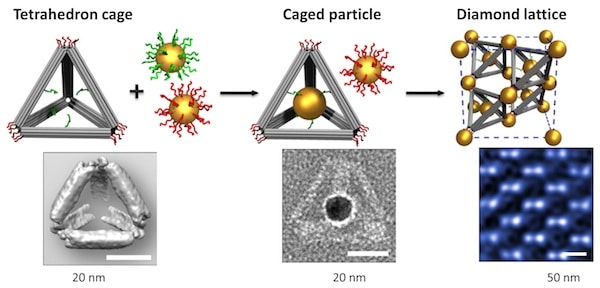
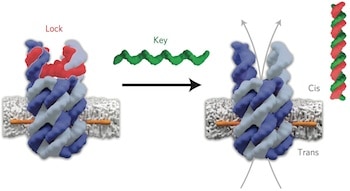
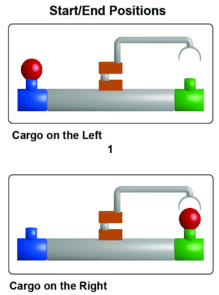
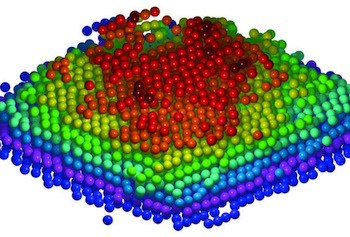
Two research teams present two different methods for using single strands of DNA to link various nanoparticles into complex 3D arrays: one using DNA hairpins for dynamic reconfiguration and the other using a DNA origami scaffold.
DNA nanotechnology provides new ways to arrange nanoparticles into crystal lattices