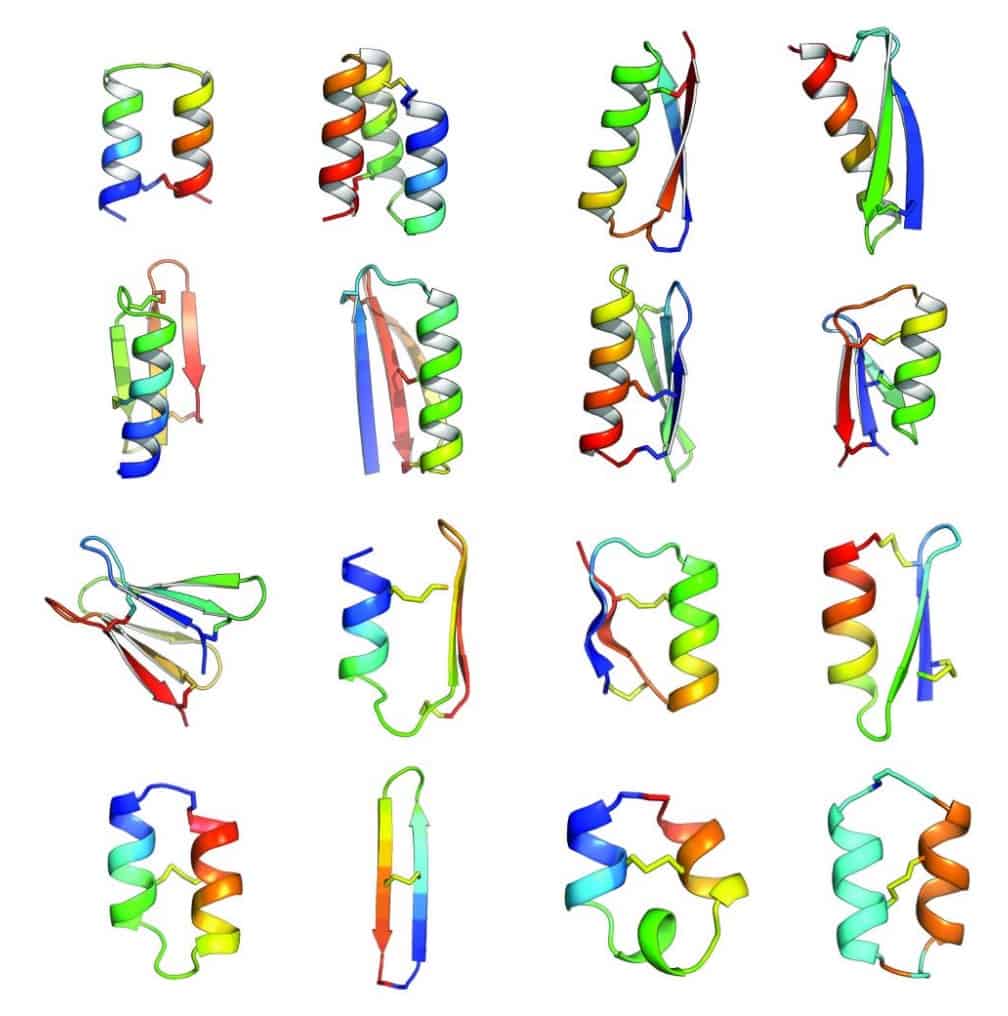
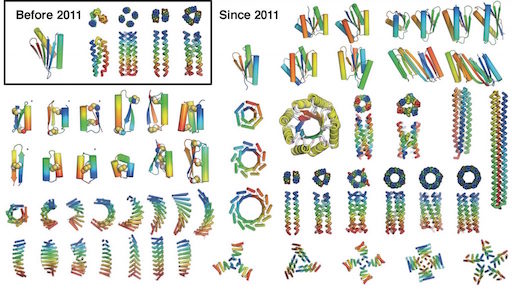
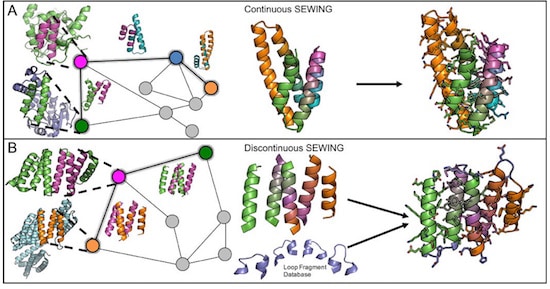
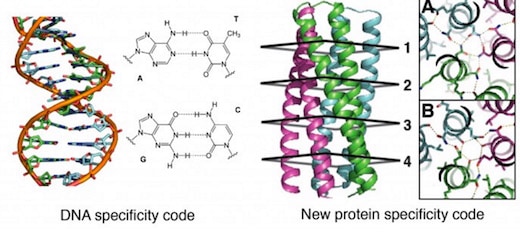
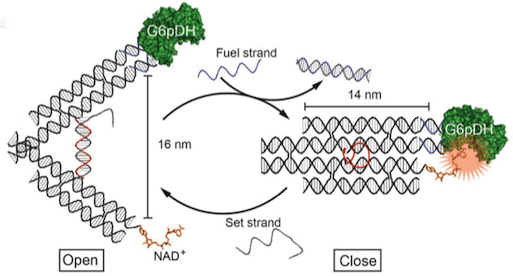
The first proposal of a path from then current technology to the ability to fabricate complex materials and devices by placing the atoms where you want them was made by Richard Feynman in 1959: “There’s Plenty of Room at the Bottom“, but see also this series “Feynman Path to Nanotechnology“. The second proposal to achieve… Continue reading Unrelated de novo enzyme replaces essential enzyme in cell
Unrelated de novo enzyme replaces essential enzyme in cell