The idea that nanorobots fabricated by atomically precise manufacturing processes are a likely part of our future, and that this is a good thing, is appearing more frequently, largely as a result of Drexler’s recent book Radical Abundance.
The idea that nanorobots fabricated by atomically precise manufacturing processes are a likely part of our future, and that this is a good thing, is appearing more frequently, largely as a result of Drexler’s recent book Radical Abundance.
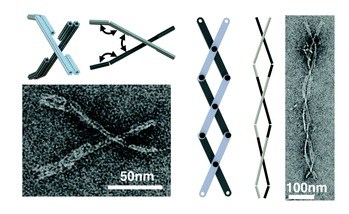
An overview of three decades of progress in DNA nanotechnology emphasizes bringing programmed motion to DNA nanostructures, including efforts to incorporate design principles from macroscopic mechanical engineering.
Scaffolded DNA origami is combined with hinges of single- or double-stranded DNA to built simple machines parts that have been combined to program simple to complex motions.
One example is presented of how well the meme is spreading that nanotechnology will evolve toward atomically precise manufacturing that will in turn bring forth a world of abundance.
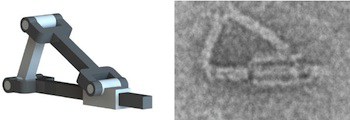
Combinations of different types of DNA nanorobots, implementing different logic gates, work together to tag a specific type of cell in a living cockroach depending on the presence or absence of two protein signals.
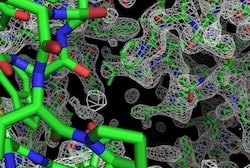
New software makes it possible to generate 3D structures of proteins without artificially incorporating metal atoms in the proteins, making it possible to study many molecular machines using data that could not previously be analyzed.
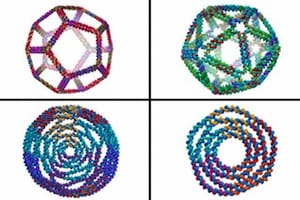
A more general computational framework predicts the structures of 2D and 3D-curved DNA nanostructures impossible to predict using previously available computational methods. May lead to 3D-printing DNA nanostructures?

A new strategy to form bonds between carbon atoms opens the way to a wide variety of molecular architectures that had been difficult or impossible to access using previous methods.
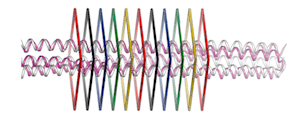
Advances in the de novo design of coiled-coil proteins made by two different research groups proceeding by two different routes demonstrate that the range of protein nanostructures potentially available for various molecular machine systems is significantly larger than the range of such structures already exploited by natural selection.

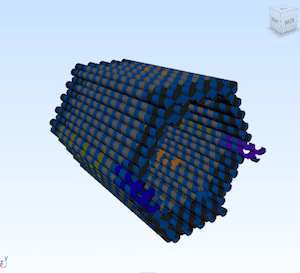
A general framework is presented for using 32-nucleotide DNA bricks to build large two-dimensional crystals up to 80 nm thick and incorporating sophisticated three-dimensional features.